 |
PDBsum entry 4j5h

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase/hydrolase substrate
|
PDB id
|
|

|

|
4j5h
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase/hydrolase substrate
|
 |
|
Title:
|
 |
Crystal structure of b. Thuringiensis aiia mutant f107w with n- decanoyl-l-homoserine bound at the active site
|
|
Structure:
|
 |
N-acyl homoserine lactonase. Chain: a. Synonym: ahl-lactonase, homoserine lactone lactonase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Bacillus thuringiensis. Organism_taxid: 1428. Gene: aiia. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.45Å
|
R-factor:
|
0.122
|
R-free:
|
0.151
|
|
|
Authors:
|
 |
C.F.Liu,D.Liu,J.Momb,P.W.Thomas,A.Lajoie,G.A.Petsko,W.Fast,D.Ringe
|
|
Key ref:
|
 |
C.F.Liu
et al.
(2013).
A phenylalanine clamp controls substrate specificity in the quorum-quenching metallo-γ-lactonase from Bacillus thuringiensis.
Biochemistry,
52,
1603-1610.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
08-Feb-13
|
Release date:
|
26-Jun-13
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
A3FJ64
(AHLL_BACTU) -
N-acyl homoserine lactonase from Bacillus thuringiensis
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
250 a.a.
250 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.1.81
- quorum-quenching N-acyl-homoserine lactonase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an N-acyl-L-homoserine lactone + H2O = an N-acyl-L-homoserine + H+
|
 |
 |
 |
 |
 |
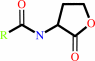 N-acyl-L-homoserine lactone
N-acyl-L-homoserine lactone
|
+
|
H2O
|
=
|
N-acyl-L-homoserine
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
52:1603-1610
(2013)
|
|
PubMed id:
|
|
|
|

|
| |
|
A phenylalanine clamp controls substrate specificity in the quorum-quenching metallo-γ-lactonase from Bacillus thuringiensis.
|
|
C.F.Liu,
D.Liu,
J.Momb,
P.W.Thomas,
A.Lajoie,
G.A.Petsko,
W.Fast,
D.Ringe.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Autoinducer inactivator A (AiiA) is a metal-dependent N-acyl homoserine lactone
hydrolase that displays broad substrate specificity but shows a preference for
substrates with long N-acyl substitutions. Previously, crystal structures of
AiiA in complex with the ring-opened product N-hexanoyl-l-homoserine revealed
binding interactions near the metal center but did not identify a binding pocket
for the N-acyl chains of longer substrates. Here we report the crystal structure
of an AiiA mutant, F107W, determined in the presence and absence of
N-decanoyl-l-homoserine. F107 is located in a hydrophobic cavity adjacent to the
previously identified ligand binding pocket, and the F107W mutation results in
the formation of an unexpected interaction with the ring-opened product.
Notably, the structure reveals a previously unidentified hydrophobic binding
pocket for the substrate's N-acyl chain. Two aromatic residues, F64 and F68,
form a hydrophobic clamp, centered around the seventh carbon in the
product-bound structure's decanoyl chain, making an interaction that would also
be available for longer substrates, but not for shorter substrates. Steady-state
kinetics using substrates of various lengths with AiiA bearing mutations at the
hydrophobic clamp, including insertion of a redox-sensitive cysteine pair,
confirms the importance of this hydrophobic feature for substrate preference.
Identifying the specificity determinants of AiiA will aid the development of
more selective quorum-quenching enzymes as tools and as potential therapeutics.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

