 |
PDBsum entry 4hpp
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.1.11
- glutamate--putrescine ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
putrescine + L-glutamate + ATP = gamma-L-glutamylputrescine + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
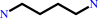 putrescine
putrescine
|
+
|
L-glutamate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 ATP
ATP
|
=
|
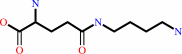 gamma-L-glutamylputrescine
gamma-L-glutamylputrescine
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
51:10121-10123
(2012)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure and activity of PA5508, a hexameric glutamine synthetase homologue.
|
|
J.E.Ladner,
V.Atanasova,
Z.Dolezelova,
J.F.Parsons.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structure of PA5508 from Pseudomonas aeruginosa, a glutamine synthetase (GS)
homologue, has been determined at 2.5 Å. Surprisingly, PA5508 forms single
hexameric rings rather than the stacked double rings that are characteristic of
GS. The C-terminal helical thong motif that links GS rings is present in PA5508;
however, it is folded back toward the core of its own polypeptide, preventing it
from interacting with a second ring. Interestingly, PA5508 displays a clear
preference for aromatic amine substrates. Unique aspects of the structure
illustrate how the enzyme is able to catalyze reactions involving bulky amines
rather than ammonia.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

