 |
PDBsum entry 4fjs

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4fjs
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of ureidoglycolate dehydrogenase enzyme in apo form
|
|
Structure:
|
 |
Ureidoglycolate dehydrogenase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 316385. Strain: dh10b. Gene: alld, b0517, ecdh10b_0473, glxb8, jw0505, ylbc. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.13Å
|
R-factor:
|
0.223
|
R-free:
|
0.240
|
|
|
Authors:
|
 |
M.I.Kim,I.Shin,J.Lee,S.Rhee
|
|
Key ref:
|
 |
M.I.Kim
et al.
(2012).
Structural and functional insights into (S)-ureidoglycolate dehydrogenase, a metabolic branch point enzyme in nitrogen utilization.
Plos One,
7,
e52066.
PubMed id:
|
 |
|
Date:
|
 |
|
12-Jun-12
|
Release date:
|
16-Jan-13
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chain A:
E.C.1.1.1.154
- ureidoglycolate dehydrogenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP Catabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
(S)-ureidoglycolate + NAD+ = N-carbamoyl-2-oxoglycine + NADH + H+
|
|
2.
|
(S)-ureidoglycolate + NADP+ = N-carbamoyl-2-oxoglycine + NADPH + H+
|
|
 |
 |
 |
 |
 |
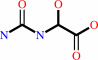 (S)-ureidoglycolate
(S)-ureidoglycolate
|
+
|
 NAD(+)
NAD(+)
|
=
|
N-carbamoyl-2-oxoglycine
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 (S)-ureidoglycolate
(S)-ureidoglycolate
|
+
|
 NADP(+)
NADP(+)
|
=
|
N-carbamoyl-2-oxoglycine
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Plos One
7:e52066
(2012)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural and functional insights into (S)-ureidoglycolate dehydrogenase, a metabolic branch point enzyme in nitrogen utilization.
|
|
M.I.Kim,
I.Shin,
S.Cho,
J.Lee,
S.Rhee.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Nitrogen metabolism is one of essential processes in living organisms. The
catabolic pathways of nitrogenous compounds play a pivotal role in the storage
and recovery of nitrogen. In Escherichia coli, two different, interconnecting
metabolic routes drive nitrogen utilization through purine degradation
metabolites. The enzyme (S)-ureidoglycolate dehydrogenase (AllD), which is a
member of l-sulfolactate dehydrogenase-like family, converts
(S)-ureidoglycolate, a key intermediate in the purine degradation pathway, to
oxalurate in an NAD(P)-dependent manner. Therefore, AllD is a metabolic
branch-point enzyme for nitrogen metabolism in E. coli. Here, we report crystal
structures of AllD in its apo form, in a binary complex with NADH cofactor, and
in a ternary complex with NADH and glyoxylate, a possible spontaneous
degradation product of oxalurate. Structural analyses revealed that NADH in an
extended conformation is bound to an NADH-binding fold with three distinct
domains that differ from those of the canonical NADH-binding fold. We also
characterized ligand-induced structural changes, as well as the binding mode of
glyoxylate, in the active site near the NADH nicotinamide ring. Based on
structural and kinetic analyses, we concluded that AllD selectively utilizes
NAD(+) as a cofactor, and further propose that His116 acts as a general
catalytic base and that a hydride transfer is possible on the B-face of the
nicotinamide ring of the cofactor. Other residues conserved in the active sites
of this novel l-sulfolactate dehydrogenase-like family also play essential roles
in catalysis.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

