 |
PDBsum entry 4f8b

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4f8b
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of the covalent thioimide intermediate of unimodular nitrile reductase quef
|
|
Structure:
|
 |
NADPH-dependent 7-cyano-7-deazaguanine reductase. Chain: a, b, c, d, e. Synonym: quef, 7-cyano-7-carbaguanine reductase, NADPH-dependent nitrile oxidoreductase, preq(0) reductase. Engineered: yes
|
|
Source:
|
 |
Bacillus subtilis subsp. Subtilis. Organism_taxid: 224308. Strain: 168. Gene: bsu13750, quef, ykvm. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.50Å
|
R-factor:
|
0.193
|
R-free:
|
0.272
|
|
|
Authors:
|
 |
B.Stec,M.A.Swairjo
|
|
Key ref:
|
 |
V.M.Chikwana
et al.
(2012).
Structural basis of biological nitrile reduction.
J Biol Chem,
287,
30560-30570.
PubMed id:
|
 |
|
Date:
|
 |
|
17-May-12
|
Release date:
|
11-Jul-12
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O31678
(QUEF_BACSU) -
NADPH-dependent 7-cyano-7-deazaguanine reductase from Bacillus subtilis (strain 168)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
165 a.a.
139 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.7.1.13
- preQ1 synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
7-aminomethyl-7-carbaguanine + 2 NADP+ = 7-cyano-7-deazaguanine + 2 NADPH + 3 H+
|
 |
 |
 |
 |
 |
7-aminomethyl-7-carbaguanine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 2
×
NADP(+)
2
×
NADP(+)
|
=
|
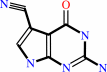 7-cyano-7-deazaguanine
7-cyano-7-deazaguanine
|
+
|
 2
×
NADPH
2
×
NADPH
|
+
|
3
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Biol Chem
287:30560-30570
(2012)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis of biological nitrile reduction.
|
|
V.M.Chikwana,
B.Stec,
B.W.Lee,
V.de Crécy-Lagard,
D.Iwata-Reuyl,
M.A.Swairjo.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

