 |
PDBsum entry 4xjs

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase, transferase
|
PDB id
|
|

|

|
4xjs
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase, transferase
|
 |
|
Title:
|
 |
Human cd38 complexed with inhibitor 1 [6-fluoro-2-methyl-4-[(2,3,6- trichlorobenzyl)amino]quinoline-8-carboxamide]
|
|
Structure:
|
 |
Adp-ribosyl cyclase/cyclic adp-ribose hydrolase 1. Chain: a. Synonym: 2'-phospho-adp-ribosyl cyclase,2'-phospho-adp-ribosyl cyclase/2'-phospho-cyclic-adp-ribose transferase,2'-phospho-cyclic- adp-ribose transferase,adp-ribosyl cyclase 1,adprc 1,cyclic adp- ribose hydrolase 1,cadpr hydrolase 1,t10. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: cd38. Expressed in: pichia. Expression_system_taxid: 4919
|
|
Resolution:
|
 |
|
2.80Å
|
R-factor:
|
0.190
|
R-free:
|
0.263
|
|
|
Authors:
|
 |
L.M.Shewchuk,D.Deaton,E.Stewart
|
|
Key ref:
|
 |
J.D.Becherer
et al.
(2015).
Discovery of 4-Amino-8-quinoline Carboxamides as Novel, Submicromolar Inhibitors of NAD-Hydrolyzing Enzyme CD38.
J Med Chem,
58,
7021-7056.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
09-Jan-15
|
Release date:
|
26-Aug-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P28907
(CD38_HUMAN) -
ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
300 a.a.
239 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 4 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.4.99.20
- 2'-phospho-ADP-ribosyl cyclase/2'-phospho-cyclic-ADP-ribose transferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
nicotinate + NADP+ = nicotinate-adenine dinucleotide phosphate + nicotinamide
|
 |
 |
 |
 |
 |
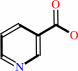 nicotinate
nicotinate
|
+
|
 NADP(+)
NADP(+)
|
=
|
nicotinate-adenine dinucleotide phosphate
|
+
|
 nicotinamide
nicotinamide
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.2.2.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.2.2.6
- ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
NAD+ + H2O = ADP-D-ribose + nicotinamide + H+
|
 |
 |
 |
 |
 |
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
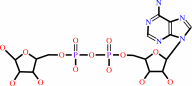 ADP-D-ribose
ADP-D-ribose
|
+
|
 nicotinamide
nicotinamide
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Med Chem
58:7021-7056
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Discovery of 4-Amino-8-quinoline Carboxamides as Novel, Submicromolar Inhibitors of NAD-Hydrolyzing Enzyme CD38.
|
|
J.D.Becherer,
E.E.Boros,
T.Y.Carpenter,
D.J.Cowan,
D.N.Deaton,
C.D.Haffner,
M.R.Jeune,
I.W.Kaldor,
J.C.Poole,
F.Preugschat,
T.R.Rheault,
C.A.Schulte,
B.G.Shearer,
T.W.Shearer,
L.M.Shewchuk,
T.L.Smalley,
E.L.Stewart,
J.D.Stuart,
J.C.Ulrich.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Starting from the micromolar 8-quinoline carboxamide high-throughput screening
hit 1a, a systematic exploration of the structure-activity relationships (SAR)
of the 4-, 6-, and 8-substituents of the quinoline ring resulted in the
identification of approximately 10-100-fold more potent human CD38 inhibitors.
Several of these molecules also exhibited pharmacokinetic parameters suitable
for in vivo animal studies, including low clearances and decent oral
bioavailability. Two of these CD38 inhibitors, 1ah and 1ai, were shown to
elevate NAD tissue levels in liver and muscle in a diet-induced obese (DIO)
C57BL/6 mouse model. These inhibitor tool compounds will enable further
biological studies of the CD38 enzyme as well as the investigation of the
therapeutic implications of NAD enhancement in disease models of abnormally low
NAD.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

