 |
PDBsum entry 4q5o

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4q5o
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.11.55
- ectoine hydroxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-ectoine + 2-oxoglutarate + O2 = 5-hydroxyectoine + succinate + CO2
|
 |
 |
 |
 |
 |
L-ectoine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
+
|
 O2
O2
|
=
|
5-hydroxyectoine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 succinate
succinate
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Ascorbate; Fe(2+)
|
 |
 |
 |
 |
 |
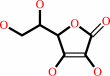 Ascorbate
Ascorbate
|
Fe(2+)
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
289:29570-29583
(2014)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the ectoine hydroxylase, a snapshot of the active site.
|
|
A.Höppner,
N.Widderich,
M.Lenders,
E.Bremer,
S.H.Smits.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Ectoine and its derivative 5-hydroxyectoine are compatible solutes that are
widely synthesized by bacteria to cope physiologically with osmotic stress. They
also serve as chemical chaperones and maintain the functionality of
macromolecules. 5-Hydroxyectoine is produced from ectoine through a
stereo-specific hydroxylation, an enzymatic reaction catalyzed by the ectoine
hydroxylase (EctD). The EctD protein is a member of the non-heme-containing
iron(II) and 2-oxoglutarate-dependent dioxygenase superfamily and is
evolutionarily well conserved. We studied the ectoine hydroxylase from the
cold-adapted marine ultra-microbacterium Sphingopyxis alaskensis (Sa) and found
that the purified SaEctD protein is a homodimer in solution. We determined the
SaEctD crystal structure in its apo-form, complexed with the iron catalyst, and
in a form that contained iron, the co-substrate 2-oxoglutarate, and the reaction
product of EctD, 5-hydroxyectoine. The iron and 2-oxoglutarate ligands are bound
within the EctD active site in a fashion similar to that found in other members
of the dioxygenase superfamily. 5-Hydroxyectoine, however, is coordinated by
EctD in manner different from that found in high affinity solute receptor
proteins operating in conjunction with microbial import systems for ectoines.
Our crystallographic analysis provides a detailed view into the active site of
the ectoine hydroxylase and exposes an intricate network of interactions between
the enzyme and its ligands that collectively ensure the hydroxylation of the
ectoine substrate in a position- and stereo-specific manner.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

