 |
PDBsum entry 4py2

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Ligase/ligase inhibitor
|
PDB id
|
|

|

|
4py2
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.10
- methionine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Met) + L-methionine + ATP = L-methionyl-tRNA(Met) + AMP + diphosphate
|
 |
 |
 |
 |
 |
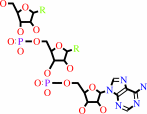 tRNA(Met)
tRNA(Met)
|
+
|
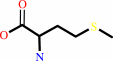 L-methionine
L-methionine
|
+
|
 ATP
ATP
|
=
|
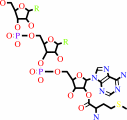 L-methionyl-tRNA(Met)
L-methionyl-tRNA(Met)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Plos One
11:e0160350
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
Brucella melitensis Methionyl-tRNA-Synthetase (MetRS), a Potential Drug Target for Brucellosis.
|
|
K.K.Ojo,
R.M.Ranade,
Z.Zhang,
D.M.Dranow,
J.B.Myers,
R.Choi,
S.Nakazawa Hewitt,
T.E.Edwards,
D.R.Davies,
D.Lorimer,
S.M.Boyle,
L.K.Barrett,
F.S.Buckner,
E.Fan,
W.C.Van Voorhis.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
We investigated Brucella melitensis methionyl-tRNA-synthetase (BmMetRS) with
molecular, structural and phenotypic methods to learn if BmMetRS is a promising
target for brucellosis drug development. Recombinant BmMetRS was expressed,
purified from wild type Brucella melitensis biovar Abortus 2308 strain ATCC/CRP
#DD-156 and screened by a thermal melt assay against a focused library of one
hundred previously classified methionyl-tRNA-synthetase inhibitors of the blood
stage form of Trypanosoma brucei. Three compounds showed appreciable shift of
denaturation temperature and were selected for further studies on inhibition of
the recombinant enzyme activity and cell viability against wild type B.
melitensis strain 16M. BmMetRS protein complexed with these three inhibitors
resolved into three-dimensional crystal structures and was analyzed. All three
selected methionyl-tRNA-synthetase compounds inhibit recombinant BmMetRS
enzymatic functions in an aminoacylation assay at varying concentrations.
Furthermore, growth inhibition of B. melitensis strain 16M by the compounds was
shown. Inhibitor-BmMetRS crystal structure models were used to illustrate the
molecular basis of the enzyme inhibition. Our current data suggests that BmMetRS
is a promising target for brucellosis drug development. However, further studies
are needed to optimize lead compound potency, efficacy and safety as well as
determine the pharmacokinetics, optimal dosage, and duration for effective
treatment.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

