 |
PDBsum entry 4poc
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.4.2.3.3
- methylglyoxal synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
dihydroxyacetone phosphate = methylglyoxal + phosphate
|
 |
 |
 |
 |
 |
 dihydroxyacetone phosphate
dihydroxyacetone phosphate
|
=
|
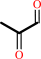 methylglyoxal
methylglyoxal
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.5.3.1.1
- triose-phosphate isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-glyceraldehyde 3-phosphate = dihydroxyacetone phosphate
|
 |
 |
 |
 |
 |
D-glyceraldehyde 3-phosphate
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
=
|
 dihydroxyacetone phosphate
dihydroxyacetone phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochim Biophys Acta
1852:61-69
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Triosephosphate isomerase I170V alters catalytic site, enhances stability and induces pathology in a Drosophila model of TPI deficiency.
|
|
B.P.Roland,
C.G.Amrich,
C.J.Kammerer,
K.A.Stuchul,
S.B.Larsen,
S.Rode,
A.A.Aslam,
A.Heroux,
R.Wetzel,
A.P.VanDemark,
M.J.Palladino.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Triosephosphate isomerase (TPI) is a glycolytic enzyme which homodimerizes for
full catalytic activity. Mutations of the TPI gene elicit a disease known as TPI
Deficiency, a glycolytic enzymopathy noted for its unique severity of
neurological symptoms. Evidence suggests that TPI Deficiency pathogenesis may be
due to conformational changes of the protein, likely affecting dimerization and
protein stability. In this report, we genetically and physically characterize a
human disease-associated TPI mutation caused by an I170V substitution. Human
TPI(I170V) elicits behavioral abnormalities in Drosophila. An examination of
hTPI(I170V) enzyme kinetics revealed this substitution reduced catalytic
turnover, while assessments of thermal stability demonstrated an increase in
enzyme stability. The crystal structure of the homodimeric I170V mutant reveals
changes in the geometry of critical residues within the catalytic pocket.
Collectively these data reveal new observations of the structural and kinetic
determinants of TPI Deficiency pathology, providing new insights into disease
pathogenesis.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

