 |
PDBsum entry 4hr0

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4hr0
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.17.4.1
- ribonucleoside-diphosphate reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 2'-deoxyribonucleoside 5'-diphosphate + [thioredoxin]-disulfide + H2O = a ribonucleoside 5'-diphosphate + [thioredoxin]-dithiol
|
 |
 |
 |
 |
 |
 2'-deoxyribonucleoside diphosphate
2'-deoxyribonucleoside diphosphate
|
+
|
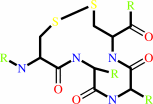 thioredoxin disulfide
thioredoxin disulfide
|
+
|
H(2)O
|
=
|
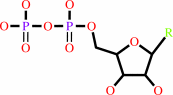 ribonucleoside diphosphate
ribonucleoside diphosphate
|
+
|
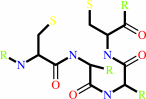 thioredoxin
thioredoxin
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe(3+) or adenosylcob(III)alamin or Mn(2+)
|
 |
 |
 |
 |
 |
Fe(3+)
|
or
|
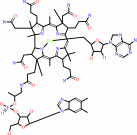 adenosylcob(III)alamin
adenosylcob(III)alamin
|
or
|
Mn(2+)
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
110:17189-17194
(2013)
|
|
PubMed id:
|
|
|
|

|
| |
|
Direct observation of structurally encoded metal discrimination and ether bond formation in a heterodinuclear metalloprotein.
|
|
J.J.Griese,
K.Roos,
N.Cox,
H.S.Shafaat,
R.M.Branca,
J.Lehtiö,
A.Gräslund,
W.Lubitz,
P.E.Siegbahn,
M.Högbom.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Although metallocofactors are ubiquitous in enzyme catalysis, how metal binding
specificity arises remains poorly understood, especially in the case of metals
with similar primary ligand preferences such as manganese and iron. The
biochemical selection of manganese over iron presents a particularly intricate
problem because manganese is generally present in cells at a lower concentration
than iron, while also having a lower predicted complex stability according to
the Irving-Williams series (Mn(II) < Fe(II) < Ni(II) < Co(II) <
Cu(II) > Zn(II)). Here we show that a heterodinuclear Mn/Fe cofactor with the
same primary protein ligands in both metal sites self-assembles from Mn(II) and
Fe(II) in vitro, thus diverging from the Irving-Williams series without
requiring auxiliary factors such as metallochaperones. Crystallographic,
spectroscopic, and computational data demonstrate that one of the two metal
sites preferentially binds Fe(II) over Mn(II) as expected, whereas the other
site is nonspecific, binding equal amounts of both metals in the absence of
oxygen. Oxygen exposure results in further accumulation of the Mn/Fe cofactor,
indicating that cofactor assembly is at least a two-step process governed by
both the intrinsic metal specificity of the protein scaffold and additional
effects exerted during oxygen binding or activation. We further show that the
mixed-metal cofactor catalyzes a two-electron oxidation of the protein scaffold,
yielding a tyrosine-valine ether cross-link. Theoretical modeling of the
reaction by density functional theory suggests a multistep mechanism including a
valyl radical intermediate.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

