 |
PDBsum entry 4gac

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4gac
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chains A, B:
E.C.1.1.1.19
- glucuronate reductase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Mammalian Ascorbic Acid Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-gulonate + NADP+ = aldehydo-D-glucuronate + NADPH + H+
|
 |
 |
 |
 |
 |
L-gulonate
Bound ligand (Het Group name = )
matches with 52.94% similarity
|
+
|
 NADP(+)
NADP(+)
|
=
|
aldehydo-D-glucuronate
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B:
E.C.1.1.1.2
- alcohol dehydrogenase (NADP(+)).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a primary alcohol + NADP+ = an aldehyde + NADPH + H+
|
 |
 |
 |
 |
 |
primary alcohol
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
+
|
 NADP(+)
NADP(+)
|
=
|
 aldehyde
aldehyde
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains A, B:
E.C.1.1.1.20
- glucuronolactone reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-gulono-1,4-lactone + NADP+ = D-glucurono-3,6-lactone + NADPH + H+
|
 |
 |
 |
 |
 |
L-gulono-1,4-lactone
Bound ligand (Het Group name = )
matches with 56.25% similarity
|
+
|
 NADP(+)
NADP(+)
|
=
|
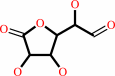 D-glucurono-3,6-lactone
D-glucurono-3,6-lactone
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
Chains A, B:
E.C.1.1.1.33
- Transferred entry: 1.1.1.2.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(R)-mevalonate + NADP+ = (R)-mevaldate + H+ + NADPH
|
 |
 |
 |
 |
 |
(R)-mevalonate
Bound ligand (Het Group name = )
matches with 76.92% similarity
|
+
|
 NADP(+)
NADP(+)
|
=
|
(R)-mevaldate
|
+
|
H(+)
|
+
|
 NADPH
NADPH
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
Chains A, B:
E.C.1.1.1.372
- D/L-glyceraldehyde reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
glycerol + NADP+ = L-glyceraldehyde + NADPH + H+
|
|
2.
|
glycerol + NADP+ = D-glyceraldehyde + NADPH + H+
|
|
 |
 |
 |
 |
 |
glycerol
Bound ligand (Het Group name = )
matches with 66.67% similarity
|
+
|
 NADP(+)
NADP(+)
|
=
|
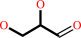 L-glyceraldehyde
L-glyceraldehyde
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
glycerol
Bound ligand (Het Group name = )
matches with 66.67% similarity
|
+
|
 NADP(+)
NADP(+)
|
=
|
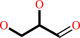 D-glyceraldehyde
D-glyceraldehyde
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 6:
|
 |
Chains A, B:
E.C.1.1.1.54
- allyl-alcohol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
allyl alcohol + NADP+ = acrolein + NADPH + H+
|
 |
 |
 |
 |
 |
allyl alcohol
Bound ligand (Het Group name = )
matches with 60.00% similarity
|
+
|
 NADP(+)
NADP(+)
|
=
|
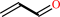 acrolein
acrolein
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr Sect F Struct Biol Cryst Commun
68:1271-1274
(2012)
|
|
PubMed id:
|
|
|
|

|
| |
|
High-resolution structure of AKR1a4 in the apo form and its interaction with ligands.
|
|
F.Faucher,
Z.Jia.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

