 |
PDBsum entry 4g8t
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of a glucarate dehydratase related protein, from actinobacillus succinogenes, target efi-502312, with sodium and sulfate bound, ordered loop
|
|
Structure:
|
 |
Glucarate dehydratase. Chain: a, b, c, d. Engineered: yes
|
|
Source:
|
 |
Actinobacillus succinogenes. Organism_taxid: 339671. Strain: 130z. Gene: asuc_1847. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.70Å
|
R-factor:
|
0.154
|
R-free:
|
0.179
|
|
|
Authors:
|
 |
M.W.Vetting,R.Toro,R.Bhosle,N.F.Al Obaidi,L.L.Morisco,S.R.Wasserman, S.Sojitra,E.Washington,A.Scott Glenn,S.Chowdhury,B.Evans,J.Hammonds, B.Hillerich,J.Love,R.D.Seidel,H.J.Imker,J.A.Gerlt,S.C.Almo,Enzyme Function Initiative (Efi)
|
|
Key ref:
|
 |
M.W.Vetting
et al.
Crystal structure of a glucarate dehydratase related protein, From actinobacillus succinogenes, Target efi-502312, With sodium and sulfate bound, Ordered lo.
To be published,
.
|
 |
|
Date:
|
 |
|
23-Jul-12
|
Release date:
|
15-Aug-12
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
A6VQF1
(A6VQF1_ACTSZ) -
glucarate dehydratase from Actinobacillus succinogenes (strain ATCC 55618 / DSM 22257 / CCUG 43843 / 130Z)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
442 a.a.
441 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.40
- glucarate dehydratase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-glucarate = 5-dehydro-4-deoxy-D-glucarate + H2O
|
 |
 |
 |
 |
 |
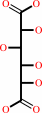 D-glucarate
D-glucarate
|
=
|
5-dehydro-4-deoxy-D-glucarate
Bound ligand (Het Group name = )
matches with 46.15% similarity
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

