 |
PDBsum entry 4dql

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4dql
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.14.14.1
- unspecific monooxygenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an organic molecule + reduced [NADPH--hemoprotein reductase] + O2 = an alcohol + oxidized [NADPH--hemoprotein reductase] + H2O + H+
|
 |
 |
 |
 |
 |
organic molecule
|
+
|
reduced [NADPH--hemoprotein reductase]
|
+
|
 O2
O2
|
=
|
 alcohol
alcohol
|
+
|
oxidized [NADPH--hemoprotein reductase]
|
+
|
H2O
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme-thiolate
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.6.2.4
- NADPH--hemoprotein reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 oxidized [cytochrome P450] + NADPH = 2 reduced [cytochrome P450] + NADP+ + H+
|
 |
 |
 |
 |
 |
2
×
oxidized [cytochrome P450]
|
+
|
 NADPH
NADPH
|
=
|
2
×
reduced [cytochrome P450]
|
+
|
 NADP(+)
NADP(+)
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD; FMN
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
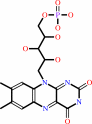 FMN
FMN
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Febs J
279:1694-1706
(2012)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crystal structure of the FAD/NADPH-binding domain of flavocytochrome P450 BM3.
|
|
M.G.Joyce,
I.S.Ekanem,
O.Roitel,
A.J.Dunford,
R.Neeli,
H.M.Girvan,
G.J.Baker,
R.A.Curtis,
A.W.Munro,
D.Leys.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

