 |
PDBsum entry 4d9p

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase/transferase inhibitor
|
PDB id
|
|

|

|
4d9p
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.5.1.15
- dihydropteroate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Folate Biosynthesis (late stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(7,8-dihydropterin-6-yl)methyl diphosphate + 4-aminobenzoate = 7,8- dihydropteroate + diphosphate
|
 |
 |
 |
 |
 |
(7,8-dihydropterin-6-yl)methyl diphosphate
|
+
|
4-aminobenzoate
Bound ligand (Het Group name = )
matches with 42.86% similarity
|
=
|
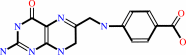 7,8- dihydropteroate
7,8- dihydropteroate
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Chemmedchem
7:861-870
(2012)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure-based design of novel pyrimido[4,5-c]pyridazine derivatives as dihydropteroate synthase inhibitors with increased affinity.
|
|
Y.Zhao,
D.Hammoudeh,
M.K.Yun,
J.Qi,
S.W.White,
R.E.Lee.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

