 |
PDBsum entry 4aop

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4aop
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.8.1.2
- assimilatory sulfite reductase (NADPH).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
hydrogen sulfide + 3 NADP+ + 3 H2O = sulfite + 3 NADPH + 4 H+
|
 |
 |
 |
 |
 |
hydrogen sulfide
|
+
|
 3
×
NADP(+)
3
×
NADP(+)
|
+
|
3
×
H2O
|
=
|
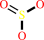 sulfite
sulfite
|
+
|
 3
×
NADPH
3
×
NADPH
|
+
|
4
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD; FMN; Iron-sulfur; Siroheme
|
 |
 |
 |
 |
 |
 FAD
FAD
|
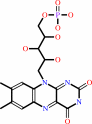 FMN
FMN
|
 Iron-sulfur
Iron-sulfur
|
Siroheme
Bound ligand (Het Group name =
SRM)
matches with 96.88% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
36:12101-12119
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of the siroheme- and Fe4S4-containing active center of sulfite reductase in different states of oxidation: heme activation via reduction-gated exogenous ligand exchange.
|
|
B.R.Crane,
L.M.Siegel,
E.D.Getzoff.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The active center of the Escherichia coli sulfite reductase hemoprotein (SiRHP)
is exquisitely designed to catalyze the six-electron reductions of sulfite to
sulfide and nitrite to ammonia. Refined high-resolution crystallographic
structures of oxidized, two-electron reduced, and intermediately reduced states
of SiRHP, monitored by single-crystal electron paramagnetic resonance (EPR)
spectroscopy, reveal that a bridging cysteine thiolate supplied by the protein
always covalently links the siroheme (iron isobacteriochlorin) to the Fe4S4
cluster, facilitating their ability to transfer electrons to substrate. The
reduction potential and reactivity of the cluster are tuned by association with
the siroheme, accessibility to solvent, and hydrogen bonds supplied by the
protein loops containing the four cluster-ligating cysteines. The distorted
conformation of the siroheme recognized by the protein potentially destabilizes
the electronic conjugation of the isobacteriochlorin ring and produces axial
configurations for some propionate side chains that promote interactions with
exogenous ligands and active-site residues. An extensive hydrogen-bond network
of positively charged side chains, ordered water molecules, and siroheme
carboxylates coordinates, polarizes, and influences the protonation state of
anionic ligands. In the oxidized (siroheme Fe3+, Fe4S42+) SiRHP crystal
structure, the high density of positive charges in the binding pocket is
stabilized by the siroheme's sixth axial ligand-an exogenous phosphate anion.
Binding assays with H32PO42- demonstrate that oxidized SiRHP binds phosphate in
solution with a dissociation constant of 14 microM at pH 7.7, suggesting that
phosphate anions play an important role in stabilizing and sequestering the
active-site of the oxidized enzyme in vivo. Reduction of the cofactors couples
changes in siroheme iron coordination geometry to changes in active-site protein
conformation, leading to phosphate release both in the crystal and in solution.
An intermediately reduced enzyme, where the siroheme is mainly ferrous (+2) and
the cluster cubane is mainly oxidized (+2), appears to have the lowest affinity
for phosphate in the crystal. Reduction-gated release of phosphate from the
substrate-binding site may explain the 10(5)-fold increase in rates of ligand
association that accompany reduction of SiRHP.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Shirodkar,
S.Reed,
M.Romine,
and
D.Saffarini
(2011).
The octahaem SirA catalyses dissimilatory sulfite reduction in Shewanella oneidensis MR-1.
|
| |
Environ Microbiol,
13,
108-115.
|
 |
|
|
|
|
 |
B.C.Tripathy,
I.Sherameti,
and
R.Oelmüller
(2010).
Siroheme: an essential component for life on earth.
|
| |
Plant Signal Behav,
5,
14-20.
|
 |
|
|
|
|
 |
K.Sekine,
Y.Sakakibara,
T.Hase,
and
N.Sato
(2009).
A novel variant of ferredoxin-dependent sulfite reductase having preferred substrate specificity for nitrite in the unicellular red alga Cyanidioschyzon merolae.
|
| |
Biochem J,
423,
91-98.
|
 |
|
|
|
|
 |
V.Daskalakis,
and
C.Varotsis
(2009).
Binding and Docking Interactions of NO, CO and O(2) in Heme Proteins as Probed by Density Functional Theory.
|
| |
Int J Mol Sci,
10,
4137-4156.
|
 |
|
|
|
|
 |
J.Zeng,
M.Wang,
X.Zhang,
Y.Wang,
C.Ai,
J.Liu,
and
G.Qiu
(2008).
Expression, purification and characterization of the sulfite reductase hemo-subunit, SiR-HP, from Acidithiobacillus ferrooxidans.
|
| |
Biotechnol Lett,
30,
1239-1244.
|
 |
|
|
|
|
 |
R.Pinto,
J.S.Harrison,
T.Hsu,
W.R.Jacobs,
and
T.S.Leyh
(2007).
Sulfite reduction in mycobacteria.
|
| |
J Bacteriol,
189,
6714-6722.
|
 |
|
|
|
|
 |
S.Cai,
T.K.h.Shokhireva,
D.L.Lichtenberger,
and
F.A.Walker
(2006).
NMR and EPR studies of chloroiron(III) tetraphenyl-chlorin and its complexes with imidazoles and pyridines of widely differing basicities.
|
| |
Inorg Chem,
45,
3519-3531.
|
 |
|
|
|
|
 |
M.Hirasawa,
M.Nakayama,
S.K.Kim,
T.Hase,
and
D.B.Knaff
(2005).
Chemical modification studies of tryptophan, arginine and lysine residues in maize chloroplast ferredoxin:sulfite oxidoreductase.
|
| |
Photosynth Res,
86,
325-336.
|
 |
|
|
|
|
 |
M.A.Wallace,
L.L.Liou,
J.Martins,
M.H.Clement,
S.Bailey,
V.D.Longo,
J.S.Valentine,
and
E.B.Gralla
(2004).
Superoxide inhibits 4Fe-4S cluster enzymes involved in amino acid biosynthesis. Cross-compartment protection by CuZn-superoxide dismutase.
|
| |
J Biol Chem,
279,
32055-32062.
|
 |
|
|
|
|
 |
I.Curdt,
B.B.Singh,
M.Jakoby,
W.Hachtel,
and
H.Böhme
(2000).
Identification of amino acid residues of nitrite reductase from Anabaena sp. PCC 7120 involved in ferredoxin binding.
|
| |
Biochim Biophys Acta,
1543,
60-68.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

