 |
PDBsum entry 3w2m

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase/oxidoreductase inhibitor
|
PDB id
|
|

|

|
3w2m
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase/oxidoreductase inhibitor
|
 |
|
Title:
|
 |
Structure of trypanosoma cruzi dihydroorotate dehydrogenase in complex with mii-3-183
|
|
Structure:
|
 |
Dihydroorotate dehydrogenase (fumarate). Chain: a, b. Synonym: dhod, dhodase, dhodehase, dihydroorotate oxidase. Engineered: yes
|
|
Source:
|
 |
Trypanosoma cruzi. Organism_taxid: 353153. Strain: cl brener. Gene: pyrd. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.58Å
|
R-factor:
|
0.139
|
R-free:
|
0.169
|
|
|
Authors:
|
 |
D.K.Inaoka,M.Iida,T.Tabuchi,N.Lee,S.Matsuoka,T.Shiba,K.Sakamoto, S.Suzuki,E.O.Balogun,T.Nara,T.Aoki,M.Inoue,T.Honma,A.Tanaka, S.Harada,K.Kita
|
|
Key ref:
|
 |
D.K.Inaoka
et al.
Structure of trypanosoma cruzi dihydroorotate dehydro in complex with mii-3-183..
To be published,
.
|
 |
|
Date:
|
 |
|
30-Nov-12
|
Release date:
|
04-Dec-13
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q4D3W2
(PYRD_TRYCC) -
Dihydroorotate dehydrogenase (fumarate) from Trypanosoma cruzi (strain CL Brener)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
314 a.a.
313 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.3.98.1
- dihydroorotate oxidase (fumarate).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-dihydroorotate + fumarate = orotate + succinate
|
 |
 |
 |
 |
 |
(S)-dihydroorotate
Bound ligand (Het Group name = )
matches with 47.83% similarity
|
+
|
fumarate
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
=
|
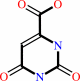 orotate
orotate
|
+
|
 succinate
succinate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FMN
|
 |
 |
 |
 |
 |
FMN
Bound ligand (Het Group name =
FMN)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

