 |
PDBsum entry 3v8e
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of the yeast nicotinamidase pnc1p bound to the inhibitor nicotinaldehyde
|
|
Structure:
|
 |
Nicotinamidase. Chain: a, b, c, d, e, f, g. Synonym: nicotine deamidase, namase. Engineered: yes
|
|
Source:
|
 |
Saccharomyces cerevisiae. Baker's yeast. Organism_taxid: 559292. Strain: atcc 204508 / s288c. Gene: pnc1, ygl037c. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.71Å
|
R-factor:
|
0.182
|
R-free:
|
0.203
|
|
|
Authors:
|
 |
K.A.Hoadley,B.C.Smith,J.M.Denu,J.L.Keck
|
|
Key ref:
|
 |
B.C.Smith
et al.
(2012).
Structural and kinetic isotope effect studies of nicotinamidase (Pnc1) from Saccharomyces cerevisiae.
Biochemistry,
51,
243-256.
PubMed id:
|
 |
|
Date:
|
 |
|
22-Dec-11
|
Release date:
|
25-Jan-12
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P53184
(PNC1_YEAST) -
Nicotinamidase from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
216 a.a.
216 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.1.19
- nicotinamidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
nicotinamide + H2O = nicotinate + NH4+
|
 |
 |
 |
 |
 |
 nicotinamide
nicotinamide
|
+
|
H2O
|
=
|
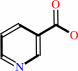 nicotinate
nicotinate
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
51:243-256
(2012)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural and kinetic isotope effect studies of nicotinamidase (Pnc1) from Saccharomyces cerevisiae.
|
|
B.C.Smith,
M.A.Anderson,
K.A.Hoadley,
J.L.Keck,
W.W.Cleland,
J.M.Denu.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

