 |
PDBsum entry 3uvh

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transport protein
|
PDB id
|
|

|

|
3uvh
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transport protein
|
 |
|
Title:
|
 |
Crystal structure analysis of e81m mutant of human clic1
|
|
Structure:
|
 |
Chloride intracellular channel protein 1. Chain: a, b. Synonym: chloride channel abp, nuclear chloride ion channel 27, ncc27, regulatory nuclear chloride ion channel protein, hrncc. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: clic1, g6, ncc27. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.84Å
|
R-factor:
|
0.204
|
R-free:
|
0.258
|
|
|
Authors:
|
 |
S.Fanucchi,I.A.Achilonu,M.A.Fernandes,D.A.Legg-E'Silva,S.H.Stoychev, H.W.Dirr
|
|
Key ref:
|
 |
D.A.Legg-E'Silva
et al.
Crystal structure analysis of e81m mutant of human cl.
To be published,
.
|
 |
|
Date:
|
 |
|
30-Nov-11
|
Release date:
|
14-Dec-11
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O00299
(CLIC1_HUMAN) -
Chloride intracellular channel protein 1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
241 a.a.
231 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.8.-.-
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.8.5.1
- glutathione dehydrogenase (ascorbate).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-dehydroascorbate + 2 glutathione = glutathione disulfide + L-ascorbate
|
 |
 |
 |
 |
 |
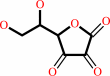 L-dehydroascorbate
L-dehydroascorbate
|
+
|
 2
×
glutathione
2
×
glutathione
|
=
|
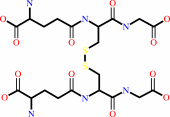 glutathione disulfide
glutathione disulfide
|
+
|
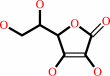 L-ascorbate
L-ascorbate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

