 |
PDBsum entry 3twb
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of gluconate dehydratase (target efi-501679) from salmonella enterica subsp. Enterica serovar enteritidis str. P125109 complexed with magnesium and gluconic acid
|
|
Structure:
|
 |
Putative dehydratase. Chain: a, b, c, d, e. Engineered: yes
|
|
Source:
|
 |
Salmonella enterica subsp. Enterica serovar enteritidis. Organism_taxid: 550537. Strain: p125109. Gene: sen1436. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.76Å
|
R-factor:
|
0.171
|
R-free:
|
0.206
|
|
|
Authors:
|
 |
Y.Patskovsky,R.Toro,R.Bhosle,B.Hillerich,R.D.Seidel,E.Washington, A.Scott Glenn,S.Chowdhury,B.Evans,J.Hammonds,W.D.Zencheck,H.J.Imker, J.A.Gerlt,S.C.Almo,Enzyme Function Initiative (Efi)
|
|
Key ref:
|
 |
Y.Patskovsky
et al.
Crystal structure of gluconate dehydratase from salmo enterica p125109.
To be published,
.
|
 |
|
Date:
|
 |
|
21-Sep-11
|
Release date:
|
26-Oct-11
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
B5R541
(DGD_SALEP) -
D-galactonate dehydratase family member SEN1436 from Salmonella enteritidis PT4 (strain P125109)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
419 a.a.
415 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.39
- gluconate dehydratase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-gluconate = 2-dehydro-3-deoxy-D-gluconate + H2O
|
 |
 |
 |
 |
 |
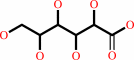 D-gluconate
D-gluconate
|
=
|
2-dehydro-3-deoxy-D-gluconate
Bound ligand (Het Group name = )
matches with 92.31% similarity
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

