 |
PDBsum entry 3rv6

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Isomerase/isomerase inhibitor
|
PDB id
|
|

|

|
3rv6
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase/isomerase inhibitor
|
 |
|
Title:
|
 |
Structure of a m. Tuberculosis salicylate synthase, mbti, in complex with an inhibitor with phenyl r-group
|
|
Structure:
|
 |
Isochorismate synthase/isochorismate-pyruvate lyase mbti. Chain: a, b. Synonym: mycobactin synthase protein i, salicylate synthase mbti. Engineered: yes
|
|
Source:
|
 |
Mycobacterium tuberculosis. Organism_taxid: 1773. Strain: h37rv. Gene: rv2386c. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.04Å
|
R-factor:
|
0.172
|
R-free:
|
0.205
|
|
|
Authors:
|
 |
G.Chi,E.M.M.Bulloch,A.Manos-Turvey,R.J.Payne,J.S.Lott,Tb Structural Genomics Consortium (Tbsgc)
|
|
Key ref:
|
 |
G.Chi
et al.
(2012).
Implications of binding mode and active site flexibility for inhibitor potency against the salicylate synthase from Mycobacterium tuberculosis.
Biochemistry,
51,
4868-4879.
PubMed id:
|
 |
|
Date:
|
 |
|
06-May-11
|
Release date:
|
09-May-12
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P9WFX1
(MBTI_MYCTU) -
Salicylate synthase from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
450 a.a.
417 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.4.2.99.21
- isochorismate lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
isochorismate = salicylate + pyruvate
|
 |
 |
 |
 |
 |
isochorismate
Bound ligand (Het Group name = )
matches with 72.73% similarity
|
=
|
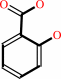 salicylate
salicylate
|
+
|
 pyruvate
pyruvate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.5.4.4.2
- isochorismate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
chorismate = isochorismate
|
 |
 |
 |
 |
 |
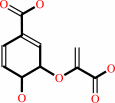 chorismate
chorismate
|
=
|
isochorismate
Bound ligand (Het Group name = )
matches with 72.73% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mg(2+)
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.5.4.99.5
- chorismate mutase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
chorismate = prephenate
|
 |
 |
 |
 |
 |
chorismate
Bound ligand (Het Group name = )
matches with 65.22% similarity
|
=
|
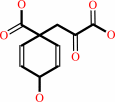 prephenate
prephenate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
51:4868-4879
(2012)
|
|
PubMed id:
|
|
|
|

|
| |
|
Implications of binding mode and active site flexibility for inhibitor potency against the salicylate synthase from Mycobacterium tuberculosis.
|
|
G.Chi,
A.Manos-Turvey,
P.D.O'Connor,
J.M.Johnston,
G.L.Evans,
E.N.Baker,
R.J.Payne,
J.S.Lott,
E.M.Bulloch.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

