 |
PDBsum entry 3r6c

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase/transferase inhibitor
|
PDB id
|
|

|

|
3r6c
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.18
- anthranilate phosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Tryptophan Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-(5-phospho-beta-D-ribosyl)anthranilate + diphosphate = 5-phospho-alpha- D-ribose 1-diphosphate + anthranilate
|
 |
 |
 |
 |
 |
N-(5-phospho-beta-D-ribosyl)anthranilate
Bound ligand (Het Group name = )
matches with 40.91% similarity
|
+
|
 diphosphate
diphosphate
|
=
|
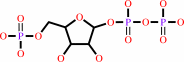 5-phospho-alpha- D-ribose 1-diphosphate
5-phospho-alpha- D-ribose 1-diphosphate
|
+
|
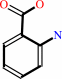 anthranilate
anthranilate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
52:1776-1787
(2013)
|
|
PubMed id:
|
|
|
|

|
| |
|
The substrate capture mechanism of Mycobacterium tuberculosis anthranilate phosphoribosyltransferase provides a mode for inhibition.
|
|
A.Castell,
F.L.Short,
G.L.Evans,
T.V.Cookson,
E.M.Bulloch,
D.D.Joseph,
C.E.Lee,
E.J.Parker,
E.N.Baker,
J.S.Lott.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Anthranilate phosphoribosyltransferase (AnPRT, EC 2.4.2.18) is a homodimeric
enzyme that catalyzes the reaction between 5'-phosphoribosyl 1'-pyrophosphate
(PRPP) and anthranilate, as part of the tryptophan biosynthesis pathway. Here we
present the results of the first chemical screen for inhibitors against
Mycobacterium tuberculosis AnPRT (Mtb-AnPRT), along with crystal structures of
Mtb-AnPRT in complex with PRPP and several inhibitors. Previous work revealed
that PRPP is bound at the base of a deep cleft in Mtb-AnPRT and predicted two
anthranilate binding sites along the tunnel leading to the PRPP binding site.
Unexpectedly, the inhibitors presented here almost exclusively bound at the
entrance of the tunnel, in the presumed noncatalytic anthranilate binding site,
previously hypothesized to have a role in substrate capture. The potencies of
the inhibitors were measured, yielding Ki values of 1.5-119 μM, with the
strongest inhibition displayed by a bianthranilate compound that makes hydrogen
bond and salt bridge contacts with Mtb-AnPRT via its carboxyl groups. Our
results reveal how the substrate capture mechanism of AnPRT can be exploited to
inhibit the enzyme's activity and provide a scaffold for the design of improved
Mtb-AnPRT inhibitors that may ultimately form the basis of new antituberculosis
drugs with a novel mode of action.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

