 |
PDBsum entry 3qpm

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
3qpm
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of peroxiredoxin prx4 from pseudosciaena crocea
|
|
Structure:
|
 |
Peroxiredoxin. Chain: a, b, c, d, e. Engineered: yes
|
|
Source:
|
 |
Larimichthys crocea. Large yellow croaker. Organism_taxid: 215358. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.90Å
|
R-factor:
|
0.197
|
R-free:
|
0.220
|
|
|
Authors:
|
 |
F.M.Lian,Y.B.Teng,Y.L.Jiang,Y.X.He,Y.Chen,C.Z.Zhou
|
|
Key ref:
|
 |
F.M.Lian
et al.
The n-Terminal beta-Sheet of peroxiredoxin prx4 in th yellow croaker pseudosciaena crocea is critical for i peroxidase and anti-Bacterial activities.
To be published,
.
|
 |
|
Date:
|
 |
|
14-Feb-11
|
Release date:
|
22-Feb-12
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
H3JQV8
(H3JQV8_LARCR) -
Peroxiredoxin from Larimichthys crocea
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
232 a.a.
195 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.11.1.15
- Transferred entry: 1.11.1.24, 1.11.1.25, 1.11.1.26, 1.11.1.27, 1.11.1.28
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Peroxiredoxin
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 R'-SH + ROOH = R'-S-S-R' + H2O + ROH
|
 |
 |
 |
 |
 |
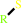 2
×
R'-SH
2
×
R'-SH
|
+
|
 ROOH
ROOH
|
=
|
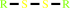 R'-S-S-R'
R'-S-S-R'
|
+
|
H(2)O
|
+
|
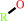 ROH
ROH
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

