 |
PDBsum entry 3pgm

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase (phosphoryl)
|
PDB id
|
|

|

|
3pgm
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.4.2.11
- phosphoglycerate mutase (2,3-diphosphoglycerate-dependent).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2R)-2-phosphoglycerate = (2R)-3-phosphoglycerate
|
 |
 |
 |
 |
 |
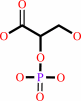 2-phospho-D-glycerate
2-phospho-D-glycerate
|
=
|
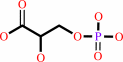 3-phospho-D-glycerate
3-phospho-D-glycerate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Philos Trans R Soc Lond B Biol Sci
293:121-130
(1981)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure and activity of phosphoglycerate mutase.
|
|
S.I.Winn,
H.C.Watson,
R.N.Harkins,
L.A.Fothergill.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structure of yeast phosphoglycerate mutase determined by X-ray
crystallographic and amino acid sequence studies has been interpreted in terms
of the chemical, kinetic and mechanistic observations made on this enzyme. There
are two histidine residues at the active site, with imidazole groups almost
parallel to each other and approximately 0.4 nm apart, positioned close to the 2
and 3 positions of the substrate. The simplest interpretation of the available
information suggests that a ping-pong type mechanism operates in which at least
one of these histidine residues participates in the phosphoryl transfer
reaction. The flexible C-terminal region also plays an important role in the
enzymic reaction.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.Wang,
L.Liu,
Z.Wei,
Z.Cheng,
Y.Lin,
and
W.Gong
(2006).
Seeing the process of histidine phosphorylation in human bisphosphoglycerate mutase.
|
| |
J Biol Chem,
281,
39642-39648.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Wang,
Z.Wei,
Q.Bian,
Z.Cheng,
M.Wan,
L.Liu,
and
W.Gong
(2004).
Crystal structure of human bisphosphoglycerate mutase.
|
| |
J Biol Chem,
279,
39132-39138.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.H.Yuen,
H.Mizuguchi,
Y.H.Lee,
P.F.Cook,
K.Uyeda,
and
C.A.Hasemann
(1999).
Crystal structure of the H256A mutant of rat testis fructose-6-phosphate,2-kinase/fructose-2,6-bisphosphatase. Fructose 6-phosphate in the active site leads to mechanisms for both mutant and wild type bisphosphatase activities.
|
| |
J Biol Chem,
274,
2176-2184.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Kostrewa,
F.Grüninger-Leitch,
A.D'Arcy,
C.Broger,
D.Mitchell,
and
A.P.van Loon
(1997).
Crystal structure of phytase from Aspergillus ficuum at 2.5 A resolution.
|
| |
Nat Struct Biol,
4,
185-190.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.J.Kurland,
and
S.J.Pilkis
(1995).
Covalent control of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase: insights into autoregulation of a bifunctional enzyme.
|
| |
Protein Sci,
4,
1023-1037.
|
 |
|
|
|
|
 |
Y.Lan,
T.Lu,
P.S.Lovett,
and
D.J.Creighton
(1995).
Evidence for a (triosephosphate isomerase-like) "catalytic loop" near the active site of glyoxalase I.
|
| |
J Biol Chem,
270,
12957-12960.
|
 |
|
|
|
|
 |
M.C.Garel,
N.Arous,
M.C.Calvin,
C.T.Craescu,
J.Rosa,
and
R.Rosa
(1994).
A recombinant bisphosphoglycerate mutase variant with acid phosphatase homology degrades 2,3-diphosphoglycerate.
|
| |
Proc Natl Acad Sci U S A,
91,
3593-3597.
|
 |
|
|
|
|
 |
G.Schneider,
Y.Lindqvist,
and
P.Vihko
(1993).
Three-dimensional structure of rat acid phosphatase.
|
| |
EMBO J,
12,
2609-2615.
|
 |
|
|
|
|
 |
L.P.Yomano,
R.K.Scopes,
and
L.O.Ingram
(1993).
Cloning, sequencing, and expression of the Zymomonas mobilis phosphoglycerate mutase gene (pgm) in Escherichia coli.
|
| |
J Bacteriol,
175,
3926-3933.
|
 |
|
|
|
|
 |
S.Tsujino,
S.Shanske,
S.Sakoda,
G.Fenichel,
and
S.DiMauro
(1993).
The molecular genetic basis of muscle phosphoglycerate mutase (PGAM) deficiency.
|
| |
Am J Hum Genet,
52,
472-477.
|
 |
|
|
|
|
 |
P.J.White,
J.Nairn,
N.C.Price,
H.G.Nimmo,
J.R.Coggins,
and
I.S.Hunter
(1992).
Phosphoglycerate mutase from Streptomyces coelicolor A3(2): purification and characterization of the enzyme and cloning and sequence analysis of the gene.
|
| |
J Bacteriol,
174,
434-440.
|
 |
|
|
|
|
 |
J.F.Bazan,
R.J.Fletterick,
and
S.J.Pilkis
(1989).
Evolution of a bifunctional enzyme: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase.
|
| |
Proc Natl Acad Sci U S A,
86,
9642-9646.
|
 |
|
|
|
|
 |
N.W.Haggarty,
B.Dunbar,
and
L.A.Fothergill
(1983).
The complete amino acid sequence of human erythrocyte diphosphoglycerate mutase.
|
| |
EMBO J,
2,
1213-1220.
|
 |
|
|
|
|
 |
H.C.Watson,
N.P.Walker,
P.J.Shaw,
T.N.Bryant,
P.L.Wendell,
L.A.Fothergill,
R.E.Perkins,
S.C.Conroy,
M.J.Dobson,
and
M.F.Tuite
(1982).
Sequence and structure of yeast phosphoglycerate kinase.
|
| |
EMBO J,
1,
1635-1640.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

