 |
PDBsum entry 3og3

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
De novo protein, lyase
|
PDB id
|
|

|

|
3og3
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
De novo protein, lyase
|
 |
|
Title:
|
 |
Crystal structure of an artificial thermostable (ba)8-barrel protein from identical half barrels
|
|
Structure:
|
 |
Imidazole glycerol phosphate synthase subunit hisf. Chain: a. Synonym: igp synthase cyclase subunit, igp synthase subunit hisf, imgp synthase subunit hisf, igps subunit hisf. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Thermotoga maritima. Organism_taxid: 2336. Gene: hisf, hisf derivative (amino acids 99-219), tm_1036. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.08Å
|
R-factor:
|
0.152
|
R-free:
|
0.196
|
|
|
Authors:
|
 |
J.M.Sperl,M.Bocola,F.List,B.Kellerer,R.Sterner
|
|
Key ref:
|
 |
J.M.Sperl
et al.
Design of an artificial thermostable (ba)8-Barrel pro from identical half barrels.
To be published,
.
|
 |
|
Date:
|
 |
|
16-Aug-10
|
Release date:
|
17-Aug-11
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9X0C6
(HIS6_THEMA) -
Imidazole glycerol phosphate synthase subunit HisF from Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
253 a.a.
242 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 98 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.3.2.10
- imidazole glycerol-phosphate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5-[(5-phospho-1-deoxy-D-ribulos-1-ylimino)methylamino]-1-(5-phospho-beta- D-ribosyl)imidazole-4-carboxamide + L-glutamine = D-erythro-1-(imidazol- 4-yl)glycerol 3-phosphate + 5-amino-1-(5-phospho-beta-D- ribosyl)imidazole-4-carboxamide + L-glutamate + H+
|
 |
 |
 |
 |
 |
5-[(5-phospho-1-deoxy-D-ribulos-1-ylimino)methylamino]-1-(5-phospho-beta- D-ribosyl)imidazole-4-carboxamide
|
+
|
 L-glutamine
L-glutamine
|
=
|
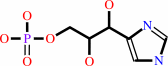 D-erythro-1-(imidazol- 4-yl)glycerol 3-phosphate
D-erythro-1-(imidazol- 4-yl)glycerol 3-phosphate
|
+
|
5-amino-1-(5-phospho-beta-D- ribosyl)imidazole-4-carboxamide
|
+
|
 L-glutamate
L-glutamate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

