 |
PDBsum entry 3o2y

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Structure-function analysis of human l-prostaglandin d synthase bound with fatty acid
|
|
Structure:
|
 |
Prostaglandin-h2 d-isomerase. Chain: a, b. Fragment: unp residues 29-190. Synonym: lipocalin-type prostaglandin-d synthase, glutathione- independent pgd synthase, prostaglandin-d2 synthase, pgd2 synthase, pgds2, pgds, beta-trace protein, cerebrin-28. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.70Å
|
R-factor:
|
0.200
|
R-free:
|
0.264
|
|
|
Authors:
|
 |
Y.Zhou,N.Shaw,Y.Li,Y.Zhao,R.Zhang,Z.-J.Liu
|
|
Key ref:
|
 |
Y.Zhou
et al.
Structure-Function analysis of human l-Prostaglandin synthase bound with fatty acid.
To be published,
.
|
 |
|
Date:
|
 |
|
23-Jul-10
|
Release date:
|
22-Sep-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P41222
(PTGDS_HUMAN) -
Prostaglandin-H2 D-isomerase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
190 a.a.
160 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.3.99.2
- prostaglandin-D synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
prostaglandin H2 = prostaglandin D2
|
 |
 |
 |
 |
 |
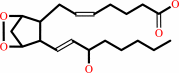 prostaglandin H2
prostaglandin H2
|
=
|
 prostaglandin D2
prostaglandin D2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Glutathione
|
 |
 |
 |
 |
 |
 Glutathione
Glutathione
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

