 |
PDBsum entry 3n1v

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase/transferase inhibitor
|
PDB id
|
|

|

|
3n1v
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.5.1.1
- dimethylallyltranstransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Terpenoid biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
isopentenyl diphosphate + dimethylallyl diphosphate = (2E)- geranyl diphosphate + diphosphate
|
 |
 |
 |
 |
 |
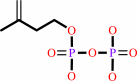 isopentenyl diphosphate
isopentenyl diphosphate
|
+
|
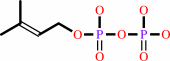 dimethylallyl diphosphate
dimethylallyl diphosphate
|
=
|
(2E)- geranyl diphosphate
|
+
|
diphosphate
Bound ligand (Het Group name = )
matches with 55.56% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.5.1.10
- (2E,6E)-farnesyl diphosphate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
isopentenyl diphosphate + (2E)-geranyl diphosphate = (2E,6E)-farnesyl diphosphate + diphosphate
|
 |
 |
 |
 |
 |
 isopentenyl diphosphate
isopentenyl diphosphate
|
+
|
(2E)-geranyl diphosphate
|
=
|
 (2E,6E)-farnesyl diphosphate
(2E,6E)-farnesyl diphosphate
|
+
|
diphosphate
Bound ligand (Het Group name = )
matches with 55.56% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Nat Chem Biol
6:660-666
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Allosteric non-bisphosphonate FPPS inhibitors identified by fragment-based discovery.
|
|
W.Jahnke,
J.M.Rondeau,
S.Cotesta,
A.Marzinzik,
X.Pellé,
M.Geiser,
A.Strauss,
M.Götte,
F.Bitsch,
R.Hemmig,
C.Henry,
S.Lehmann,
J.F.Glickman,
T.P.Roddy,
S.J.Stout,
J.R.Green.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.S.Tan,
G.Fuentes,
and
C.Verma
(2011).
A comparison of the dynamics of pantothenate synthetase from M. tuberculosis and E. coli: Computational studies.
|
| |
Proteins,
79,
1715-1727.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

