 |
PDBsum entry 3n1c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of the phosphofructokinase-2 from escherichia coli in complex with fructose-6-phosphate
|
|
Structure:
|
 |
6-phosphofructokinase isozyme 2. Chain: a, b, c, d. Synonym: phosphofructokinase-2. Engineered: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 83333. Strain: k12. Gene: b1723, jw5280, pfk2, pfkb. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.182
|
R-free:
|
0.221
|
|
|
Authors:
|
 |
H.M.Pereira,R.Cabrera,A.Caniuguir,R.C.Garratt,J.Babul
|
|
Key ref:
|
 |
R.Cabrera
et al.
(2011).
The crystal complex of phosphofructokinase-2 of Escherichia coli with fructose-6-phosphate: kinetic and structural analysis of the allosteric ATP inhibition.
J Biol Chem,
286,
5774-5783.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
15-May-10
|
Release date:
|
08-Dec-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P06999
(PFKB_ECOLI) -
ATP-dependent 6-phosphofructokinase isozyme 2 from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
309 a.a.
309 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.1.11
- 6-phosphofructokinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 6-phosphate + ATP = beta-D-fructose 1,6-bisphosphate + ADP + H+
|
 |
 |
 |
 |
 |
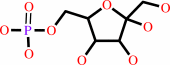 beta-D-fructose 6-phosphate
beta-D-fructose 6-phosphate
|
+
|
ATP
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
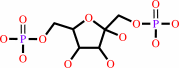 beta-D-fructose 1,6-bisphosphate
beta-D-fructose 1,6-bisphosphate
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
286:5774-5783
(2011)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crystal complex of phosphofructokinase-2 of Escherichia coli with fructose-6-phosphate: kinetic and structural analysis of the allosteric ATP inhibition.
|
|
R.Cabrera,
M.Baez,
H.M.Pereira,
A.Caniuguir,
R.C.Garratt,
J.Babul.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Substrate inhibition by ATP is a regulatory feature of the phosphofructokinases
isoenzymes from Escherichia coli (Pfk-1 and Pfk-2). Under gluconeogenic
conditions, the loss of this regulation in Pfk-2 causes substrate cycling of
fructose-6-phosphate (fructose-6-P) and futile consumption of ATP delaying
growth. In the present work, we have broached the mechanism of ATP-induced
inhibition of Pfk-2 from both structural and kinetic perspectives. The crystal
structure of Pfk-2 in complex with fructose-6-P is reported to a resolution of 2
Å. The comparison of this structure with the previously reported inhibited
form of the enzyme suggests a negative interplay between fructose-6-P binding
and allosteric binding of MgATP. Initial velocity experiments show a linear
increase of the apparent K(0.5) for fructose-6-P and a decrease in the apparent
k(cat) as a function of MgATP concentration. These effects occur simultaneously
with the induction of a sigmoidal kinetic behavior (n(H) of approximately 2).
Differences and resemblances in the patterns of fructose-6-P binding and the
mechanism of inhibition are discussed for Pfk-1 and Pfk-2, as an example of
evolutionary convergence, because these enzymes do not share a common ancestor.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

