 |
PDBsum entry 3mpb

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.3.1.15
- D-lyxose ketol-isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-lyxose = D-xylulose
|
 |
 |
 |
 |
 |
D-lyxose
Bound ligand (Het Group name = )
matches with 83.33% similarity
|
=
|
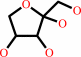 D-xylulose
D-xylulose
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Mol Biol
401:866-881
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure-based annotation of a novel sugar isomerase from the pathogenic E. coli O157:H7.
|
|
L.M.van Staalduinen,
C.S.Park,
S.J.Yeom,
M.A.Adams-Cioaba,
D.K.Oh,
Z.Jia.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Prokaryotes can use a variety of sugars as carbon sources in order to provide a
selective survival advantage. The gene z5688 found in the pathogenic E. coli
O157:H7 encodes a 'hypothetical' protein of unknown function. Sequence analysis
identified the gene product as a putative member of the cupin superfamily of
proteins, but no other functional information was known. We have determined the
crystal structure of the Z5688 protein at 1.6 A resolution and identified the
protein as a novel E. coli sugar isomerase (EcSI) through overall fold analysis
and secondary structure matching. Extensive substrate screening revealed that
EcSI is capable of acting on d-lyxose and d-mannose. The complex structure of
EcSI with fructose allowed the identification of key active site residues, and
mutagenesis confirmed their importance. The structure of EcSI also suggested a
novel mechanism for substrate binding and product release in a cupin sugar
isomerase. Supplementation of a non-pathogenic E. coli strain with EcSI enabled
cell growth on the rare pentose d-lyxose.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Marles-Wright,
and
R.J.Lewis
(2011).
The structure of a D-lyxose isomerase from the σ(B) regulon of Bacillus subtilis.
|
| |
Proteins,
79,
2015-2019.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

