 |
PDBsum entry 3mfd

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
The structure of the beta-lactamase superfamily domain of d-alanyl-d- alanine carboxypeptidase from bacillus subtilis
|
|
Structure:
|
 |
D-alanyl-d-alanine carboxypeptidase dacb. Chain: a, b. Fragment: beta-lactamase domain residues 27-358. Synonym: dd-carboxypeptidase, dd-peptidase, penicillin-binding protein 5 , Pbp-5 . Engineered: yes
|
|
Source:
|
 |
Bacillus subtilis. Organism_taxid: 1423. Strain: 168. Gene: bsu23190, dacb, dacc. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.75Å
|
R-factor:
|
0.166
|
R-free:
|
0.190
|
|
|
Authors:
|
 |
M.E.Cuff,E.Rakowski,K.Buck,A.Joachimiak,Midwest Center For Structural Genomics (Mcsg)
|
|
Key ref:
|
 |
M.E.Cuff
et al.
The structure of the beta-Lactamase superfamily domai d-Alanyl-D-Alanine carboxypeptidase from bacillus sub.
To be published,
.
|
 |
|
Date:
|
 |
|
01-Apr-10
|
Release date:
|
19-May-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.3.4.16.4
- serine-type D-Ala-D-Ala carboxypeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-alanyl-D-alanine + H2O = 2 D-alanine
|
 |
 |
 |
 |
 |
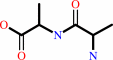
|
+
|
|
=
|
2
×
Bound ligand (Het Group name = )
matches with 42.00% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
|

