 |
PDBsum entry 3k5t

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
3k5t
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of human diamine oxidase in space group c2221
|
|
Structure:
|
 |
Diamine oxidase. Chain: a. Synonym: amiloride-sensitive amine oxidase [copper-containing], dao, amiloride-binding protein, abp, histaminase, kidney amine oxidase, kao. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: abp1, aoc1, dao1. Expressed in: drosophila melanogaster. Expression_system_taxid: 7227. Expression_system_cell_line: drosophila schneider 2 (s2)
|
|
Resolution:
|
 |
|
2.11Å
|
R-factor:
|
0.239
|
R-free:
|
0.290
|
|
|
Authors:
|
 |
A.P.Mcgrath,J.M.Guss
|
|
Key ref:
|
 |
A.P.McGrath
et al.
(2010).
A new crystal form of human diamine oxidase.
Acta Crystallogr Sect F Struct Biol Cryst Commun,
66,
137-142.
PubMed id:
|
 |
|
Date:
|
 |
|
08-Oct-09
|
Release date:
|
09-Feb-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P19801
(AOC1_HUMAN) -
Amiloride-sensitive amine oxidase [copper-containing] from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
751 a.a.
712 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 3 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.4.3.22
- diamine oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
histamine + O2 + H2O = imidazole-4-acetaldehyde + H2O2 + NH4+
|
 |
 |
 |
 |
 |
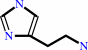 histamine
histamine
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
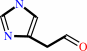 imidazole-4-acetaldehyde
imidazole-4-acetaldehyde
|
+
|
H2O2
Bound ligand (Het Group name = )
matches with 46.67% similarity
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Acta Crystallogr Sect F Struct Biol Cryst Commun
66:137-142
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
A new crystal form of human diamine oxidase.
|
|
A.P.McGrath,
K.M.Hilmer,
C.A.Collyer,
D.M.Dooley,
J.M.Guss.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Copper amine oxidases (CAOs) are ubiquitous in nature and catalyse the oxidative
deamination of primary amines to the corresponding aldehydes. Humans have three
viable CAO genes (AOC1-3). AOC1 encodes human diamine oxidase (hDAO), which is
the frontline enzyme for histamine metabolism. hDAO is unique among CAOs in that
it has a distinct substrate preference for diamines. The structure of hDAO in
space group P2(1)2(1)2(1) with two molecules in the asymmetric unit has recently
been reported. Here, the structure of hDAO refined to 2.1 A resolution in space
group C222(1) with one molecule in the asymmetric unit is reported.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

