 |
PDBsum entry 3i6t

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Crystal structure of muconate cycloisomerase from jannaschia sp.
|
|
Structure:
|
 |
Muconate cycloisomerase. Chain: a, b, c, d. Engineered: yes
|
|
Source:
|
 |
Jannaschia sp.. Organism_taxid: 290400. Strain: ccs1. Gene: jann_1408. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.90Å
|
R-factor:
|
0.170
|
R-free:
|
0.205
|
|
|
Authors:
|
 |
J.B.Bonanno,J.Freeman,K.T.Bain,J.Do,R.Romero,J.M.Sauder,S.K.Burley, S.C.Almo,New York Sgx Research Center For Structural Genomics (Nysgxrc)
|
|
Key ref:
|
 |
J.B.Bonanno
et al.
Crystal structure of muconate cycloisomerase from jannaschia sp..
To be published,
.
|
 |
|
Date:
|
 |
|
07-Jul-09
|
Release date:
|
21-Jul-09
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q28SI7
(Q28SI7_JANSC) -
Muconate cycloisomerase from Jannaschia sp. (strain CCS1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
371 a.a.
356 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.5.1.1
- muconate cycloisomerase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Benzoate Metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-muconolactone = cis,cis-muconate + H+
|
 |
 |
 |
 |
 |
(S)-muconolactone
|
=
|
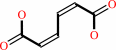 cis,cis-muconate
cis,cis-muconate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

