 |
PDBsum entry 3hgx

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.4.2.99.21
- isochorismate lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
isochorismate = salicylate + pyruvate
|
 |
 |
 |
 |
 |
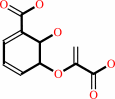 isochorismate
isochorismate
|
=
|
salicylate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
pyruvate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.5.4.99.5
- chorismate mutase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
chorismate = prephenate
|
 |
 |
 |
 |
 |
chorismate
Bound ligand (Het Group name = )
matches with 52.94% similarity
|
=
|
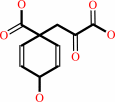 prephenate
prephenate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
48:5239-5245
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure-function analyses of isochorismate-pyruvate lyase from Pseudomonas aeruginosa suggest differing catalytic mechanisms for the two pericyclic reactions of this bifunctional enzyme.
|
|
Q.Luo,
J.Olucha,
A.L.Lamb.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The isochorismate-pyruvate lyase from Pseudomonas aeruginosa (PchB) catalyzes
two pericyclic reactions in a single active site. PchB physiologically produces
salicylate and pyruvate from isochorismate for ultimate incorporation of the
salicylate into the siderophore pyochelin. PchB also produces prephenate from
chorismate, most likely due to structural homology to the Escherchia coli
chorismate mutase. The molecular basis of catalysis among enzymatic pericyclic
reactions is a matter of debate, one view holding that catalysis may be derived
from electrostatic transition state stabilization and the opposing view that
catalysis is derived from the generation of a reactive substrate conformation.
Mutant forms of PchB were generated by site-directed mutagenesis at the site
(K42) hypothesized to be key for electrostatic transition state stabilization
(K42A, K42Q, K42E, and K42H). The loop containing K42 is mobile, and a mutant to
slow loop dynamics was also designed (A43P). Finally, a previously characterized
mutation (I87T) was also produced. Circular dichroism was used to assess the
overall effect on secondary structure as a result of the mutations, and X-ray
crystallographic structures are reported for K42A with salicylate and pyruvate
bound and for apo-I87T. The data illustrate that the active site architecture is
maintained in K42A-PchB, which indicates that differences in activity are not
caused by secondary structural changes or by differences in active site loop
conformation but rather by the chemical nature of this key residue. In contrast,
the I87T structure demonstrates considerable mobility, suggesting that loop
dynamics and conformational plasticity may be important for efficient catalysis.
Finally, the mutational effects on k(cat) provide evidence that the two
activities of PchB are not covariant and that a single hypothesis may not
provide a sufficient explanation for catalysis.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

