 |
PDBsum entry 3hdh

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
3hdh
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C:
E.C.1.1.1.35
- 3-hydroxyacyl-CoA dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a (3S)-3-hydroxyacyl-CoA + NAD+ = a 3-oxoacyl-CoA + NADH + H+
|
 |
 |
 |
 |
 |
 (3S)-3-hydroxyacyl-CoA
(3S)-3-hydroxyacyl-CoA
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
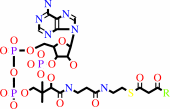 3-oxoacyl-CoA
3-oxoacyl-CoA
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
8:2010-2018
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Pig heart short chain L-3-hydroxyacyl-CoA dehydrogenase revisited: sequence analysis and crystal structure determination.
|
|
J.J.Barycki,
L.K.O'Brien,
J.J.Birktoft,
A.W.Strauss,
L.J.Banaszak.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Short chain L-3-hydroxyacyl CoA dehydrogenase (SCHAD) is a soluble dimeric
enzyme critical for oxidative metabolism of fatty acids. Its primary sequence
has been reported to be conserved across numerous tissues and species with the
notable exception of the pig heart homologue. Preliminary efforts to solve the
crystal structure of the dimeric pig heart SCHAD suggested the unprecedented
occurrence of three enzyme subunits within the asymmetric unit, a phenomenon
that was thought to have hampered refinement of the initial chain tracing. The
recently solved crystal coordinates of human heart SCHAD facilitated a molecular
replacement solution to the pig heart SCHAD data. Refinement of the model, in
conjunction with the nucleotide sequence for pig heart SCHAD determined in this
paper, has demonstrated that the previously published pig heart SCHAD sequence
was incorrect. Presented here are the corrected amino acid sequence and the high
resolution crystal structure determined for pig heart SCHAD complexed with its
NAD+ cofactor (2.8 A; R(cryst) = 22.4%, R(free) = 28.8%). In addition, the
peculiar phenomenon of a dimeric enzyme crystallizing with three subunits
contained in the asymmetric unit is described.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Arima,
A.Uesumi,
H.Mitsuzumi,
and
N.Mori
(2010).
Biochemical characterization of L-carnitine dehydrogenases from Rhizobium sp. and Xanthomonas translucens.
|
| |
Biosci Biotechnol Biochem,
74,
1237-1242.
|
 |
|
|
|
|
 |
M.Ishikawa,
D.Tsuchiya,
T.Oyama,
Y.Tsunaka,
and
K.Morikawa
(2004).
Structural basis for channelling mechanism of a fatty acid beta-oxidation multienzyme complex.
|
| |
EMBO J,
23,
2745-2754.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Vockley,
R.H.Singh,
and
D.A.Whiteman
(2002).
Diagnosis and management of defects of mitochondrial beta-oxidation.
|
| |
Curr Opin Clin Nutr Metab Care,
5,
601-609.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |
|

