 |
PDBsum entry 3h2e

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase/transferase inhibitor
|
PDB id
|
|

|

|
3h2e
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.2.5.1.15
- dihydropteroate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Folate Biosynthesis (late stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(7,8-dihydropterin-6-yl)methyl diphosphate + 4-aminobenzoate = 7,8- dihydropteroate + diphosphate
|
 |
 |
 |
 |
 |
(7,8-dihydropterin-6-yl)methyl diphosphate
|
+
|
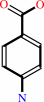 4-aminobenzoate
4-aminobenzoate
|
=
|
7,8- dihydropteroate
Bound ligand (Het Group name = )
matches with 48.15% similarity
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Med Chem
53:166-177
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural studies of pterin-based inhibitors of dihydropteroate synthase.
|
|
K.E.Hevener,
M.K.Yun,
J.Qi,
I.D.Kerr,
K.Babaoglu,
J.G.Hurdle,
K.Balakrishna,
S.W.White,
R.E.Lee.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Dihydropteroate synthase (DHPS) is a key enzyme in bacterial folate synthesis
and the target of the sulfonamide class of antibacterials. Resistance and
toxicities associated with sulfonamides have led to a decrease in their clinical
use. Compounds that bind to the pterin binding site of DHPS, as opposed to the
p-amino benzoic acid (pABA) binding site targeted by the sulfonamide agents, are
anticipated to bypass sulfonamide resistance. To identify such inhibitors and
map the pterin binding pocket, we have performed virtual screening, synthetic,
and structural studies using Bacillus anthracis DHPS. Several compounds with
inhibitory activity have been identified, and crystal structures have been
determined that show how the compounds engage the pterin site. The structural
studies identify the key binding elements and have been used to generate a
structure-activity based pharmacophore map that will facilitate the development
of the next generation of DHPS inhibitors which specifically target the pterin
site.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.W.Pemble,
P.K.Mehta,
S.Mehra,
Z.Li,
A.Nourse,
R.E.Lee,
and
S.W.White
(2010).
Crystal structure of the 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase•dihydropteroate synthase bifunctional enzyme from Francisella tularensis.
|
| |
PLoS One,
5,
e14165.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |
|

