 |
PDBsum entry 3f9m

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Human pancreatic glucokinase in complex with glucose and activator showing a mobile flap
|
|
Structure:
|
 |
Glucokinase. Chain: a. Fragment: unp residues 12-465. Synonym: hexokinase type iv, hk iv, hexokinase-4, hk4, hexokinase-d. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.50Å
|
R-factor:
|
0.191
|
R-free:
|
0.222
|
|
|
Authors:
|
 |
P.Petit,L.Gluais,A.Lagarde,L.Vuillard,J.A.Boutin,G.Ferry
|
|
Key ref:
|
 |
P.Petit
et al.
(2011).
The active conformation of human glucokinase is not altered by allosteric activators.
Acta Crystallogr D Biol Crystallogr,
67,
929-935.
PubMed id:
|
 |
|
Date:
|
 |
|
14-Nov-08
|
Release date:
|
02-Dec-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P35557
(HXK4_HUMAN) -
Hexokinase-4 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
465 a.a.
451 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 8 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.1.1
- hexokinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a D-hexose + ATP = a D-hexose 6-phosphate + ADP + H+
|
 |
 |
 |
 |
 |
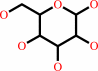 D-hexose
D-hexose
|
+
|
ATP
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
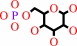 D-hexose 6-phosphate
D-hexose 6-phosphate
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Acta Crystallogr D Biol Crystallogr
67:929-935
(2011)
|
|
PubMed id:
|
|
|
|

|
| |
|
The active conformation of human glucokinase is not altered by allosteric activators.
|
|
P.Petit,
M.Antoine,
G.Ferry,
J.A.Boutin,
A.Lagarde,
L.Gluais,
R.Vincentelli,
L.Vuillard.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

