 |
PDBsum entry 3f0d

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|


 155 a.a.
155 a.a.
|
 |

|

|

|

|

|

|

|
 144 a.a.
144 a.a.
|
 |

|

|

|

|

|

|

|

 145 a.a.
145 a.a.
|
 |

|

|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
High resolution crystal structure of 2c-methyl-d-erythritol 2,4- cyclodiphosphatase synthase from burkholderia pseudomallei
|
|
Structure:
|
 |
2-c-methyl-d-erythritol 2,4-cyclodiphosphate synthase. Chain: a, b, c, d, e, f. Synonym: mecps, mecdp-synthase. Engineered: yes
|
|
Source:
|
 |
Burkholderia pseudomallei. Pseudomonas pseudomallei. Organism_taxid: 28450. Strain: 1710b. Gene: ispf, mecs, bpsl2098. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.20Å
|
R-factor:
|
0.186
|
R-free:
|
0.209
|
|
|
Authors:
|
 |
Seattle Structural Genomics Center For Infectious Disease (Ssgcid)
|
|
Key ref:
|
 |
D.W.Begley
et al.
(2011).
Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics,
12,
63-76.
PubMed id:
|
 |
|
Date:
|
 |
|
24-Oct-08
|
Release date:
|
04-Nov-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q3JRA0
(ISPF_BURP1) -
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei (strain 1710b)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
162 a.a.
155 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F:
E.C.4.6.1.12
- 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-CDP-2-C-methyl-D-erythritol 2-phosphate = 2-C-methyl-D-erythritol 2,4- cyclic diphosphate + CMP
|
 |
 |
 |
 |
 |
4-CDP-2-C-methyl-D-erythritol 2-phosphate
|
=
|
2-C-methyl-D-erythritol 2,4- cyclic diphosphate
|
+
|
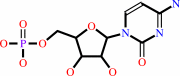 CMP
CMP
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mn(2+) or Mg(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Struct Funct Genomics
12:63-76
(2011)
|
|
PubMed id:
|
|
|
|

|
| |
|
Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
|
|
D.W.Begley,
R.C.Hartley,
D.R.Davies,
T.E.Edwards,
J.T.Leonard,
J.Abendroth,
C.A.Burris,
J.Bhandari,
P.J.Myler,
B.L.Staker,
L.J.Stewart.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.A.Bell,
K.L.Ho,
and
R.Farid
(2012).
Significant reduction in errors associated with nonbonded contacts in protein crystal structures: automated all-atom refinement with PrimeX.
|
| |
Acta Crystallogr D Biol Crystallogr,
68,
935-952.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |
| |

