 |
PDBsum entry 3euf

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.3
- uridine phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
uridine + phosphate = alpha-D-ribose 1-phosphate + uracil
|
 |
 |
 |
 |
 |
uridine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
phosphate
Bound ligand (Het Group name = )
matches with 60.87% similarity
|
=
|
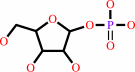 alpha-D-ribose 1-phosphate
alpha-D-ribose 1-phosphate
|
+
|
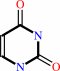 uracil
uracil
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Bmc Struct Biol
9:14
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Implications of the structure of human uridine phosphorylase 1 on the development of novel inhibitors for improving the therapeutic window of fluoropyrimidine chemotherapy.
|
|
T.P.Roosild,
S.Castronovo,
M.Fabbiani,
G.Pizzorno.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: Uridine phosphorylase (UPP) is a key enzyme of pyrimidine salvage
pathways, catalyzing the reversible phosphorolysis of ribosides of uracil to
nucleobases and ribose 1-phosphate. It is also a critical enzyme in the
activation of pyrimidine-based chemotherapeutic compounds such a 5-fluorouracil
(5-FU) and its prodrug capecitabine. Additionally, an elevated level of this
enzyme in certain tumours is believed to contribute to the selectivity of such
drugs. However, the clinical effectiveness of these fluoropyrimidine
antimetabolites is hampered by their toxicity to normal tissue. In response to
this limitation, specific inhibitors of UPP, such as 5-benzylacyclouridine
(BAU), have been developed and investigated for their ability to modulate the
cytotoxic side effects of 5-FU and its derivatives, so as to increase the
therapeutic index of these agents. RESULTS: In this report we present the high
resolution structures of human uridine phosphorylase 1 (hUPP1) in ligand-free
and BAU-inhibited conformations. The structures confirm the unexpected solution
observation that the human enzyme is dimeric in contrast to the hexameric
assembly present in microbial UPPs. They also reveal in detail the mechanism by
which BAU engages the active site of the protein and subsequently disables the
enzyme by locking the protein in a closed conformation. The observed
inter-domain motion of the dimeric human enzyme is much greater than that seen
in previous UPP structures and may result from the simpler oligomeric
organization. CONCLUSION: The structural details underlying hUPP1's active site
and additional surfaces beyond these catalytic residues, which coordinate
binding of BAU and other acyclouridine analogues, suggest avenues for future
design of more potent inhibitors of this enzyme. Notably, the loop forming the
back wall of the substrate binding pocket is conformationally different and
substantially less flexible in hUPP1 than in previously studied microbial
homologues. These distinctions can be utilized to discover novel inhibitory
compounds specifically optimized for efficacy against the human enzyme as a step
toward the development of more effective chemotherapeutic regimens that can
selectively protect normal tissues with inherently lower UPP activity.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Paul,
S.E.O'Leary,
K.Rajashankar,
W.Bu,
A.Toms,
E.C.Settembre,
J.M.Sanders,
T.P.Begley,
and
S.E.Ealick
(2010).
Glycal formation in crystals of uridine phosphorylase.
|
| |
Biochemistry,
49,
3499-3509.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.T.Larson,
D.G.Mudeppa,
J.R.Gillespie,
N.Mueller,
A.J.Napuli,
J.A.Arif,
J.Ross,
T.L.Arakaki,
A.Lauricella,
G.Detitta,
J.Luft,
F.Zucker,
C.L.Verlinde,
E.Fan,
W.C.Van Voorhis,
F.S.Buckner,
P.K.Rathod,
W.G.Hol,
and
E.A.Merritt
(2010).
The crystal structure and activity of a putative trypanosomal nucleoside phosphorylase reveal it to be a homodimeric uridine phosphorylase.
|
| |
J Mol Biol,
396,
1244-1259.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.P.Roosild,
and
S.Castronovo
(2010).
Active site conformational dynamics in human uridine phosphorylase 1.
|
| |
PLoS One,
5,
e12741.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

