 |
PDBsum entry 3eca

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of escherichia coli l-asparaginase, an enzyme used in cancer therapy (elspar)
|
|
Structure:
|
 |
L-asparaginase 2. Chain: a, b, c, d. Other_details: the structure was determined for the clinical drug elspar. The amino acid sequence differed in four places from the sequence of the k12 strain of e. Coli asparaginase (v27a, n64d, s252t, t263n).
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 562
|
|
Biol. unit:
|
 |
Tetramer (from
)
|
|
Resolution:
|
 |
|
|
Authors:
|
 |
A.L.Swain,M.Jaskolski,D.Housset,J.K.M.Rao,A.Wlodawer
|
|
Key ref:
|
 |
A.L.Swain
et al.
(1993).
Crystal structure of Escherichia coli L-asparaginase, an enzyme used in cancer therapy.
Proc Natl Acad Sci U S A,
90,
1474-1478.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
02-Jul-93
|
Release date:
|
31-Oct-93
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P00805
(ASPG2_ECOLI) -
L-asparaginase 2 from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
348 a.a.
326 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 4 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.1.1
- asparaginase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-asparagine + H2O = L-aspartate + NH4+
|
 |
 |
 |
 |
 |
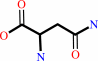 L-asparagine
L-asparagine
|
+
|
H2O
|
=
|
L-aspartate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
90:1474-1478
(1993)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of Escherichia coli L-asparaginase, an enzyme used in cancer therapy.
|
|
A.L.Swain,
M.Jaskólski,
D.Housset,
J.K.Rao,
A.Wlodawer.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of Escherichia coli asparaginase II (EC 3.5.1.1), a drug
(Elspar) used for the treatment of acute lymphoblastic leukemia, has been
determined at 2.3 A resolution by using data from a single heavy atom derivative
in combination with molecular replacement. The atomic model was refined to an R
factor of 0.143. This enzyme, active as a homotetramer with 222 symmetry,
belongs to the class of alpha/beta proteins. Each subunit has two domains with
unique topological features. On the basis of present structural evidence
consistent with previous biochemical studies, we propose locations for the
active sites between the N- and C-terminal domains belonging to different
subunits and postulate a catalytic role for Thr-89.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.N.Offman,
M.Krol,
N.Patel,
S.Krishnan,
J.Liu,
V.Saha,
and
P.A.Bates
(2011).
Rational engineering of L-asparaginase reveals importance of dual activity for cancer cell toxicity.
|
| |
Blood,
117,
1614-1621.
|
 |
|
|
|
|
 |
C.Scotti,
P.Sommi,
M.V.Pasquetto,
D.Cappelletti,
S.Stivala,
P.Mignosi,
M.Savio,
L.R.Chiarelli,
G.Valentini,
V.M.Bolanos-Garcia,
D.S.Merrell,
S.Franchini,
M.L.Verona,
C.Bolis,
E.Solcia,
R.Manca,
D.Franciotta,
A.Casasco,
P.Filipazzi,
E.Zardini,
and
V.Vannini
(2010).
Cell-cycle inhibition by Helicobacter pylori L-asparaginase.
|
| |
PLoS One,
5,
e13892.
|
 |
|
|
|
|
 |
R.S.Prakasham,
M.Hymavathi,
C.h.Subba Rao,
S.K.Arepalli,
J.Venkateswara Rao,
P.K.Kennady,
K.Nasaruddin,
J.B.Vijayakumar,
and
P.N.Sarma
(2010).
Evaluation of antineoplastic activity of extracellular asparaginase produced by isolated Bacillus circulans.
|
| |
Appl Biochem Biotechnol,
160,
72-80.
|
 |
|
|
|
|
 |
N.Patel,
S.Krishnan,
M.N.Offman,
M.Krol,
C.X.Moss,
C.Leighton,
F.W.van Delft,
M.Holland,
J.Liu,
S.Alexander,
C.Dempsey,
H.Ariffin,
M.Essink,
T.O.Eden,
C.Watts,
P.A.Bates,
and
V.Saha
(2009).
A dyad of lymphoblastic lysosomal cysteine proteases degrades the antileukemic drug L-asparaginase.
|
| |
J Clin Invest,
119,
1964-1973.
|
 |
|
|
|
|
 |
P.Dhavala,
and
A.C.Papageorgiou
(2009).
Structure of Helicobacter pyloriL-asparaginase at 1.4 A resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
1253-1261.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Masetti,
and
A.Pession
(2009).
First-line treatment of acute lymphoblastic leukemia with pegasparaginase.
|
| |
Biologics,
3,
359-368.
|
 |
|
|
|
|
 |
K.Sheppard,
J.Yuan,
M.J.Hohn,
B.Jester,
K.M.Devine,
and
D.Söll
(2008).
From one amino acid to another: tRNA-dependent amino acid biosynthesis.
|
| |
Nucleic Acids Res,
36,
1813-1825.
|
 |
|
|
|
|
 |
M.Chruszcz,
A.Wlodawer,
and
W.Minor
(2008).
Determination of protein structures--a series of fortunate events.
|
| |
Biophys J,
95,
1-9.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
O.V.Kravchenko,
Y.A.Kislitsin,
A.N.Popov,
S.V.Nikonov,
and
I.P.Kuranova
(2008).
Three-dimensional structures of L-asparaginase from Erwinia carotovora complexed with aspartate and glutamate.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
248-256.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Dhavala,
J.Krasotkina,
C.Dubreuil,
and
A.C.Papageorgiou
(2008).
Expression, purification and crystallization of Helicobacter pylori L-asparaginase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
740-742.
|
 |
|
|
|
|
 |
C.H.Fu,
and
K.M.Sakamoto
(2007).
PEG-asparaginase.
|
| |
Expert Opin Pharmacother,
8,
1977-1984.
|
 |
|
|
|
|
 |
M.K.Yun,
A.Nourse,
S.W.White,
C.O.Rock,
and
R.J.Heath
(2007).
Crystal structure and allosteric regulation of the cytoplasmic Escherichia coli L-asparaginase I.
|
| |
J Mol Biol,
369,
794-811.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Verma,
K.Kumar,
G.Kaur,
and
S.Anand
(2007).
L-asparaginase: a promising chemotherapeutic agent.
|
| |
Crit Rev Biotechnol,
27,
45-62.
|
 |
|
|
|
|
 |
H.Geckil,
S.Gencer,
B.Ates,
U.Ozer,
M.Uckun,
and
I.Yilmaz
(2006).
Effect of Vitreoscilla hemoglobin on production of a chemotherapeutic enzyme, L-asparaginase, by Pseudomonas aeruginosa.
|
| |
Biotechnol J,
1,
203-208.
|
 |
|
|
|
|
 |
J.Wang,
and
E.R.Kantrowitz
(2006).
Trapping the tetrahedral intermediate in the alkaline phosphatase reaction by substitution of the active site serine with threonine.
|
| |
Protein Sci,
15,
2395-2401.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.H.Lucas,
B.A.Ersoy,
L.A.Kueltzo,
S.B.Joshi,
D.T.Brandau,
N.Thyagarajapuram,
L.J.Peek,
and
C.R.Middaugh
(2006).
Probing protein structure and dynamics by second-derivative ultraviolet absorption analysis of cation-{pi} interactions.
|
| |
Protein Sci,
15,
2228-2243.
|
 |
|
|
|
|
 |
S.Benjwal,
S.Verma,
K.H.Röhm,
and
O.Gursky
(2006).
Monitoring protein aggregation during thermal unfolding in circular dichroism experiments.
|
| |
Protein Sci,
15,
635-639.
|
 |
|
|
|
|
 |
A.Khushoo,
Y.Pal,
and
K.J.Mukherjee
(2005).
Optimization of extracellular production of recombinant asparaginase in Escherichia coli in shake-flask and bioreactor.
|
| |
Appl Microbiol Biotechnol,
68,
189-197.
|
 |
|
|
|
|
 |
A.Werner,
K.H.Röhm,
and
H.J.Müller
(2005).
Mapping of B-cell epitopes in E. coli asparaginase II, an enzyme used in leukemia treatment.
|
| |
Biol Chem,
386,
535-540.
|
 |
|
|
|
|
 |
E.Schmitt,
M.Panvert,
S.Blanquet,
and
Y.Mechulam
(2005).
Structural basis for tRNA-dependent amidotransferase function.
|
| |
Structure,
13,
1421-1433.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.E.Wikman,
J.Krasotkina,
A.Kuchumova,
N.N.Sokolov,
and
A.C.Papageorgiou
(2005).
Crystallization and preliminary crystallographic analysis of L-asparaginase from Erwinia carotovora.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
407-409.
|
 |
|
|
|
|
 |
M.Yao,
Y.Yasutake,
H.Morita,
and
I.Tanaka
(2005).
Structure of the type I L-asparaginase from the hyperthermophilic archaeon Pyrococcus horikoshii at 2.16 angstroms resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
294-301.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Borek,
K.Michalska,
K.Brzezinski,
A.Kisiel,
J.Podkowinski,
D.T.Bonthron,
D.Krowarsch,
J.Otlewski,
and
M.Jaskolski
(2004).
Expression, purification and catalytic activity of Lupinus luteus asparagine beta-amidohydrolase and its Escherichia coli homolog.
|
| |
Eur J Biochem,
271,
3215-3226.
|
 |
|
|
|
|
 |
N.S.Murthy,
and
J.R.Knox
(2004).
Hydration of proteins: SAXS study of native and methoxy polyethyleneglycol (mPEG)-modified L-asparaginase and bovine serum albumin in mPEG solutions.
|
| |
Biopolymers,
74,
457-466.
|
 |
|
|
|
|
 |
D.C.Dieterich,
M.Landwehr,
C.Reissner,
K.H.Smalla,
K.Richter,
G.Wolf,
T.M.Böckers,
E.D.Gundelfinger,
and
M.R.Kreutz
(2003).
Gliap--a novel untypical L-asparaginase localized to rat brain astrocytes.
|
| |
J Neurochem,
85,
1117-1125.
|
 |
|
|
|
|
 |
J.Lubkowski,
M.Dauter,
K.Aghaiypour,
A.Wlodawer,
and
Z.Dauter
(2003).
Atomic resolution structure of Erwinia chrysanthemi L-asparaginase.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
84-92.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Sanches,
J.A.Barbosa,
R.T.de Oliveira,
J.Abrahão Neto,
and
I.Polikarpov
(2003).
Structural comparison of Escherichia coli L-asparaginase in two monoclinic space groups.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
416-422.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Christendat,
V.Saridakis,
Y.Kim,
P.A.Kumar,
X.Xu,
A.Semesi,
A.Joachimiak,
C.H.Arrowsmith,
and
A.M.Edwards
(2002).
The crystal structure of hypothetical protein MTH1491 from Methanobacterium thermoautotrophicum.
|
| |
Protein Sci,
11,
1409-1414.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.D.Pullinger,
R.Sowdhamini,
and
A.J.Lax
(2001).
Localization of functional domains of the mitogenic toxin of Pasteurella multocida.
|
| |
Infect Immun,
69,
7839-7850.
|
 |
|
|
|
|
 |
M.Jaskólski,
M.Kozak,
J.Lubkowski,
G.Palm,
and
A.Wlodawer
(2001).
Structures of two highly homologous bacterial L-asparaginases: a case of enantiomorphic space groups.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
369-377.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Derst,
J.Henseling,
and
K.H.Röhm
(2000).
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase activity by amino acid replacements at position 248.
|
| |
Protein Sci,
9,
2009-2017.
|
 |
|
|
|
|
 |
E.Ortlund,
M.W.Lacount,
K.Lewinski,
and
L.Lebioda
(2000).
Reactions of Pseudomonas 7A glutaminase-asparaginase with diazo analogues of glutamine and asparagine result in unexpected covalent inhibitions and suggests an unusual catalytic triad Thr-Tyr-Glu.
|
| |
Biochemistry,
39,
1199-1204.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.P.Aung,
M.Bocola,
S.Schleper,
and
K.H.Röhm
(2000).
Dynamics of a mobile loop at the active site of Escherichia coli asparaginase.
|
| |
Biochim Biophys Acta,
1481,
349-359.
|
 |
|
|
|
|
 |
L.Guo,
J.Wang,
S.Qian,
X.Yan,
R.Chen,
and
G.Meng
(2000).
Construction and structural modeling of a single-chain Fv-asparaginase fusion protein resistant to proteolysis.
|
| |
Biotechnol Bioeng,
70,
456-463.
|
 |
|
|
|
|
 |
L.Ortuño-Olea,
and
S.Durán-Vargas
(2000).
The L-asparagine operon of Rhizobium etli contains a gene encoding an atypical asparaginase.
|
| |
FEMS Microbiol Lett,
189,
177-182.
|
 |
|
|
|
|
 |
M.Paetzel,
R.E.Dalbey,
and
N.C.Strynadka
(2000).
The structure and mechanism of bacterial type I signal peptidases. A novel antibiotic target.
|
| |
Pharmacol Ther,
87,
27-49.
|
 |
|
|
|
|
 |
I.Polikarpov,
R.T.de Oliveira,
and
J.Abrahão-Neto
(1999).
Preparation and preliminary X-ray diffraction studies of a new crystal form of L-asparaginase from Escherichia coli.
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
1616-1617.
|
 |
|
|
|
|
 |
A.V.Efimov
(1997).
Structural trees for protein superfamilies.
|
| |
Proteins,
28,
241-260.
|
 |
|
|
|
|
 |
J.Lubkowski,
G.J.Palm,
G.L.Gilliland,
C.Derst,
K.H.Röhm,
and
A.Wlodawer
(1996).
Crystal structure and amino acid sequence of Wolinella succinogenes L-asparaginase.
|
| |
Eur J Biochem,
241,
201-207.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Derst,
A.Wehner,
V.Specht,
and
K.H.Röhm
(1994).
States and functions of tyrosine residues in Escherichia coli asparaginase II.
|
| |
Eur J Biochem,
224,
533-540.
|
 |
|
|
|
|
 |
G.E.Norris,
T.J.Stillman,
B.F.Anderson,
and
E.N.Baker
(1994).
The three-dimensional structure of PNGase F, a glycosylasparaginase from Flavobacterium meningosepticum.
|
| |
Structure,
2,
1049-1059.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.H.Chung,
D.R.Benson,
V.W.Cornish,
and
P.G.Schultz
(1993).
Probing the role of loop 2 in Ras function with unnatural amino acids.
|
| |
Proc Natl Acad Sci U S A,
90,
10145-10149.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

