 |
PDBsum entry 3ec7

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
3ec7
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of putative dehydrogenase from salmonella typhimurium lt2
|
|
Structure:
|
 |
Putative dehydrogenase. Chain: a, b, c, d, e, f, g, h. Engineered: yes
|
|
Source:
|
 |
Salmonella typhimurium. Organism_taxid: 99287. Strain: lt2. Expressed in: escherichia coli.
|
|
Resolution:
|
 |
|
2.15Å
|
R-factor:
|
0.179
|
R-free:
|
0.231
|
|
|
Authors:
|
 |
Y.Kim,E.Evdokimova,M.Kudritska,A.Savchenko,A.Edwards,A.Joachimiak, Midwest Center For Structural Genomics (Mcsg)
|
|
Key ref:
|
 |
Y.Kim
et al.
Crystal structure of putative dehydrogenase from salmonella typhimurium lt2.
To be published,
.
|
 |
|
Date:
|
 |
|
29-Aug-08
|
Release date:
|
23-Sep-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q8ZK57
(IOLG_SALTY) -
Inositol 2-dehydrogenase from Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
336 a.a.
336 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.18
- inositol 2-dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
myo-inositol + NAD+ = scyllo-inosose + NADH + H+
|
 |
 |
 |
 |
 |
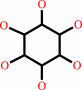 myo-inositol
myo-inositol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
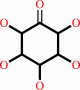 scyllo-inosose
scyllo-inosose
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

