 |
PDBsum entry 3e05

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of precorrin-6y c5,15-methyltransferase from geobacter metallireducens gs-15
|
|
Structure:
|
 |
Precorrin-6y c5,15-methyltransferase (decarboxylating). Chain: a, b, c, d, e, f, g, h. Fragment: c-terminal domain, residues 211-405. Engineered: yes
|
|
Source:
|
 |
Geobacter metallireducens. Organism_taxid: 269799. Strain: gs-15. Gene: gmet_0481. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.80Å
|
R-factor:
|
0.217
|
R-free:
|
0.262
|
|
|
Authors:
|
 |
Y.Patskovsky,U.A.Ramagopal,R.Toro,M.Dickey,S.Hu,M.Maletic,J.M.Sauder, S.K.Burley,S.C.Almo,New York Sgx Research Center For Structural Genomics (Nysgxrc)
|
|
Date:
|
 |
|
30-Jul-08
|
Release date:
|
12-Aug-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q39YF0
(Q39YF0_GEOMG) -
Cobalt-precorrin-6B C5,C15-methyltransferase and C12-decarboxylase from Geobacter metallireducens (strain ATCC 53774 / DSM 7210 / GS-15)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
405 a.a.
188 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.1.132
- precorrin-6B C(5,15)-methyltransferase (decarboxylating).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Corrin Biosynthesis (part 4)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
precorrin-6B + 2 S-adenosyl-L-methionine = precorrin-8X + 2 S-adenosyl-L- homocysteine + CO2 + 3 H+
|
 |
 |
 |
 |
 |
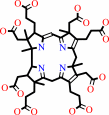 precorrin-6B
precorrin-6B
|
+
|
 2
×
S-adenosyl-L-methionine
2
×
S-adenosyl-L-methionine
|
=
|
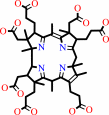 precorrin-8X
precorrin-8X
|
+
|
 2
×
S-adenosyl-L- homocysteine
2
×
S-adenosyl-L- homocysteine
|
+
|
 CO2
CO2
|
+
|
3
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

