 |
PDBsum entry 3d8h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Crystal structure of phosphoglycerate mutase from cryptosporidium parvum, cgd7_4270
|
|
Structure:
|
 |
Glycolytic phosphoglycerate mutase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Cryptosporidium parvum. Organism_taxid: 353152. Strain: iowa ii. Gene: cgd7_4270. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.01Å
|
R-factor:
|
0.233
|
R-free:
|
0.256
|
|
|
Authors:
|
 |
A.K.Wernimont,J.Lew,G.Wasney,Z.Alam,I.Kozieradzki,D.Cossar, M.Schapiro,A.Bochkarev,C.H.Arrowsmith,C.Bountra,M.Wilkstrom, A.M.Edwards,R.Hui,J.D.Artz,T.Hills,Structural Genomics Consortium (Sgc)
|
|
Key ref:
|
 |
T.Hills
et al.
(2011).
Characterization of a new phosphatase from Plasmodium.
Mol Biochem Parasitol,
179,
69-79.
PubMed id:
|
 |
|
Date:
|
 |
|
23-May-08
|
Release date:
|
15-Jul-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q5CXZ9
(Q5CXZ9_CRYPI) -
Phosphoglycerate mutase from Cryptosporidium parvum (strain Iowa II)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
249 a.a.
229 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.4.2.11
- phosphoglycerate mutase (2,3-diphosphoglycerate-dependent).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2R)-2-phosphoglycerate = (2R)-3-phosphoglycerate
|
 |
 |
 |
 |
 |
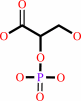 2-phospho-D-glycerate
2-phospho-D-glycerate
|
=
|
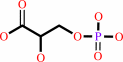 3-phospho-D-glycerate
3-phospho-D-glycerate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Mol Biochem Parasitol
179:69-79
(2011)
|
|
PubMed id:
|
|
|
|

|
| |
|
Characterization of a new phosphatase from Plasmodium.
|
|
T.Hills,
A.Srivastava,
K.Ayi,
A.K.Wernimont,
K.Kain,
A.P.Waters,
R.Hui,
J.C.Pizarro.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

