 |
PDBsum entry 3d4t

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
3d4t
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
65:229-240
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure of the periplasmic thiol-disulfide oxidoreductase SoxS from Paracoccus pantotrophus indicates a triple Trx/Grx/DsbC functionality in chemotrophic sulfur oxidation.
|
|
Y.Carius,
D.Rother,
C.G.Friedrich,
A.J.Scheidig.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The periplasmic thiol-disulfide oxidoreductase SoxS is beneficial for the
sulfur-oxidizing (Sox) phenotype of the facultative chemotrophic bacterium
Paracoccus pantotrophus and is not part of the Sox enzyme system. SoxS combines
features of thioredoxins, glutaredoxins and the thiol-disulfide oxidoreductases
of the Dsb family in structure, target specificity and reaction. The structure
of SoxS was solved in oxidized and reduced forms at 2.1 and 1.9 A resolution,
respectively. SoxS revealed high structural homology to typical cytoplasmic
bacterial thioredoxins. In contrast, SoxS contained the active-site motif
Pro-Gly-Cys-Leu-Tyr-Cys that is not present in other thioredoxins.
Interestingly, the sequence of this motif is closely related to the
Pro-Gly-Cys-Pro-Tyr-Cys sequence of some glutaredoxins and to the
Pro-Xaa-Cys-Xaa-Tyr-Cys sequences of some members of the DsbC and DsbG
subfamilies of thiol-disulfide oxidoreductases. Furthermore, the proposed
substrate of SoxS, the interprotein disulfide of SoxY, Cys110(Y)-Cys110(Y), is
structurally similar to oxidized glutathione. However, SoxS is proposed to
specifically reduce the interprotein disulfide between two SoxY subunits,
releasing a heterodimeric SoxYZ as an active part of the sulfur-oxidation cycle.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Figure 5 Stereo representation of the hydrogen-bonding network
around the thiolate of cysteine Cys13 in the reduced form of
SoxS. The main-chain trace of the protein is displayed as a loop
representation. The side chains of relevant residues are
depicted in ball-and-stick representation and coloured green,
with associated N, O and S atoms in blue, red and yellow,
respectively. Direct and water-mediated hydrogen bonds are
represented by cyan dashed lines. This figure was prepared using
PyMOL (DeLano, 2004[DeLano, W. L. (2004). The PyMOL Molecular
Graphics System. http://www.pymol.org .]).
|
 |
Figure 6.
Figure 6 Molecular-surface representation of SoxS. (a)
Representation of the molecular surface coloured according to
the electrostatic potential. The yellow asterisk indicates the
position of the redox-active Cys13. The molecular surface is
coloured according to the electrostatic potential as calculated
with the program APBS (Baker et al., 2001[Baker, N. A., Sept,
D., Joseph, S., Holst, M. J. & McCammon, J. A. (2001). Proc.
Natl Acad. Sci. USA, 98, 10037-10041.]). The molecular surface
is colour-ramped according to the electrostatic potential, with
red indicating negative potential and blue indicating positive
potential; fully saturated colours indicate a potential of 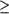 or
equal to] ±4kT/e (assuming an ionic strength of 150 mM, a
protein interior dielectric of 2 and a solvent dielectric of
78.5). The rendered surface representation was prepared with
PyMOL (DeLano, 2004[DeLano, W. L. (2004). The PyMOL Molecular
Graphics System. http://www.pymol.org .]). (b) The putative
binding cleft on the surface of SoxS. The spheres are coloured
according to the type of the underlying atom (carbon, green;
nitrogen, blue; oxygen, red; sulfur, yellow). For the putative
substrate-binding epitope the C atoms are coloured magenta. The
S atom of the redox-active cysteinyl residue Cys13 is labelled
as well as the aromatic amino-acid residues located at the
surface near the active site. or
equal to] ±4kT/e (assuming an ionic strength of 150 mM, a
protein interior dielectric of 2 and a solvent dielectric of
78.5). The rendered surface representation was prepared with
PyMOL (DeLano, 2004[DeLano, W. L. (2004). The PyMOL Molecular
Graphics System. http://www.pymol.org .]). (b) The putative
binding cleft on the surface of SoxS. The spheres are coloured
according to the type of the underlying atom (carbon, green;
nitrogen, blue; oxygen, red; sulfur, yellow). For the putative
substrate-binding epitope the C atoms are coloured magenta. The
S atom of the redox-active cysteinyl residue Cys13 is labelled
as well as the aromatic amino-acid residues located at the
surface near the active site.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2009,
65,
229-240)
copyright 2009.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

