 |
PDBsum entry 3d4m

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
3d4m
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Glutaredoxin 2 oxidized structure
|
|
Structure:
|
 |
Glutaredoxin-2, mitochondrial. Chain: a. Synonym: thioltransferase, glutathione-dependent oxidoreductase 2. Engineered: yes
|
|
Source:
|
 |
Saccharomyces cerevisiae. Yeast. Organism_taxid: 4932. Gene: grx2, ttr, ttr1, ydr513w, d9719.17. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.05Å
|
R-factor:
|
0.182
|
R-free:
|
0.219
|
|
|
Authors:
|
 |
K.F.Discola,M.A.De Oliveira,J.A.Barcena,P.Porras,B.G.Guimaraes, L.E.S.Netto
|
Key ref:

|
 |
K.F.Discola
et al.
(2009).
Structural aspects of the distinct biochemical properties of glutaredoxin 1 and glutaredoxin 2 from Saccharomyces cerevisiae.
J Mol Biol,
385,
889-901.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
14-May-08
|
Release date:
|
28-Oct-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P17695
(GLRX2_YEAST) -
Glutaredoxin-2 from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
143 a.a.
109 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.11.1.9
- glutathione peroxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 glutathione + H2O2 = glutathione disulfide + 2 H2O
|
 |
 |
 |
 |
 |
 2
×
glutathione
2
×
glutathione
|
+
|
 H2O2
H2O2
|
=
|
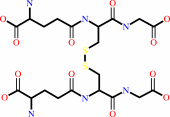 glutathione disulfide
glutathione disulfide
|
+
|
2
×
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Se(2+)
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.2.5.1.18
- glutathione transferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
RX + glutathione = an S-substituted glutathione + a halide anion + H+
|
 |
 |
 |
 |
 |
 2
×
RX
2
×
RX
|
+
|
 glutathione
glutathione
|
=
|
S-substituted glutathione
|
+
|
2
×
halide anion
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
385:889-901
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural aspects of the distinct biochemical properties of glutaredoxin 1 and glutaredoxin 2 from Saccharomyces cerevisiae.
|
|
K.F.Discola,
M.A.de Oliveira,
J.R.Rosa Cussiol,
G.Monteiro,
J.A.Bárcena,
P.Porras,
C.A.Padilla,
B.G.Guimarães,
L.E.Netto.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Glutaredoxins (Grxs) are small (9-12 kDa) heat-stable proteins that are
ubiquitously distributed. In Saccharomyces cerevisiae, seven Grx enzymes have
been identified. Two of them (yGrx1 and yGrx2) are dithiolic, possessing a
conserved Cys-Pro-Tyr-Cys motif. Here, we show that yGrx2 has a specific
activity 15 times higher than that of yGrx1, although these two oxidoreductases
share 64% identity and 85% similarity with respect to their amino acid
sequences. Further characterization of the enzymatic activities through
two-substrate kinetics analysis revealed that yGrx2 possesses a lower K(M) for
glutathione and a higher turnover than yGrx1. To better comprehend these
biochemical differences, the pK(a) of the N-terminal active-site cysteines
(Cys27) of these two proteins and of the yGrx2-C30S mutant were determined.
Since the pK(a) values of the yGrx1 and yGrx2 Cys27 residues are very similar,
these parameters cannot account for the difference observed between their
specific activities. Therefore, crystal structures of yGrx2 in the oxidized form
and with a glutathionyl mixed disulfide were determined at resolutions of 2.05
and 1.91 A, respectively. Comparisons of yGrx2 structures with the recently
determined structures of yGrx1 provided insights into their remarkable
functional divergence. We hypothesize that the substitutions of Ser23 and Gln52
in yGrx1 by Ala23 and Glu52 in yGrx2 modify the capability of the active-site
C-terminal cysteine to attack the mixed disulfide between the N-terminal
active-site cysteine and the glutathione molecule. Mutagenesis studies supported
this hypothesis. The observed structural and functional differences between
yGrx1 and yGrx2 may reflect variations in substrate specificity.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Fig. 5. Cartoon representation of the overall fold of yGrx1
and yGrx2 showing the structural alignment of yGrx1[GS]
(α-carbon atoms in violet; PDB code 2JAC),^22 yGrx2[ox]
(α-carbon atoms in gray) and yGrx2[GS] (α-carbon atoms in
cyan). The GSH molecule bonded to the yGrx2-C30S mutant is shown
in yellow.
|
 |
Figure 8.
Fig. 8. Side-chain conformation of Ser30 in the structures of
the C30S mutants of yGrx1^22 (gray) and yGrx2 (cyan) in their
glutathionylated forms. It is clear that the distances from the
Cys27 sulfur atoms (yellow) to the Ser30 oxygen atom (red) are
very different between yGrx1 and yGrx2, indicating that the
conformation of a Ser30 residue in yGrx2 is less favorable to
attack the mixed disulfide.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2009,
385,
889-901)
copyright 2009.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.R.Pedrajas,
C.A.Padilla,
B.McDonagh,
and
J.A.Bárcena
(2010).
Glutaredoxin participates in the reduction of peroxides by the mitochondrial 1-CYS peroxiredoxin in Saccharomyces cerevisiae.
|
| |
Antioxid Redox Signal,
13,
249-258.
|
 |
|
|
|
|
 |
L.Li,
N.Cheng,
K.D.Hirschi,
and
X.Wang
(2010).
Structure of Arabidopsis chloroplastic monothiol glutaredoxin AtGRXcp.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
725-732.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Smiri,
A.Chaoui,
N.Rouhier,
C.Kamel,
E.Gelhaye,
J.P.Jacquot,
and
E.El Ferjani
(2010).
Cadmium induced mitochondrial redox changes in germinating pea seed.
|
| |
Biometals,
23,
973-984.
|
 |
|
|
|
|
 |
V.E.Marquez,
D.G.Arias,
C.V.Piattoni,
C.Robello,
A.A.Iglesias,
and
S.A.Guerrero
(2010).
Cloning, expression, and characterization of a dithiol glutaredoxin from Trypanosoma cruzi.
|
| |
Antioxid Redox Signal,
12,
787-792.
|
 |
|
|
|
|
 |
M.J.Saaranen,
K.E.Salo,
M.K.Latva-Ranta,
V.L.Kinnula,
and
L.W.Ruddock
(2009).
The C-terminal active site cysteine of Escherichia coli glutaredoxin 1 determines the glutathione specificity of the second step of peptide deglutathionylation.
|
| |
Antioxid Redox Signal,
11,
1819-1828.
|
 |
|
|
|
|
 |
M.M.Gallogly,
D.W.Starke,
and
J.J.Mieyal
(2009).
Mechanistic and kinetic details of catalysis of thiol-disulfide exchange by glutaredoxins and potential mechanisms of regulation.
|
| |
Antioxid Redox Signal,
11,
1059-1081.
|
 |
|
|
|
|
 |
Z.R.Zhang,
and
S.Perrett
(2009).
Novel Glutaredoxin Activity of the Yeast Prion Protein Ure2 Reveals a Native-like Dimer within Fibrils.
|
| |
J Biol Chem,
284,
14058-14067.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

