 |
PDBsum entry 3d1v

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.1
- purine-nucleoside phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a purine D-ribonucleoside + phosphate = a purine nucleobase + alpha- D-ribose 1-phosphate
|
|
2.
|
a purine 2'-deoxy-D-ribonucleoside + phosphate = a purine nucleobase + 2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine D-ribonucleoside
|
+
|
 phosphate
phosphate
|
=
|
purine nucleobase
|
+
|
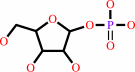 alpha- D-ribose 1-phosphate
alpha- D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine 2'-deoxy-D-ribonucleoside
|
+
|
 phosphate
phosphate
|
=
|
purine nucleobase
|
+
|
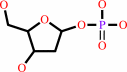 2-deoxy-alpha-D-ribose 1-phosphate
2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Arch Biochem Biophys
479:28-38
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural studies of human purine nucleoside phosphorylase: towards a new specific empirical scoring function.
|
|
L.F.Timmers,
R.A.Caceres,
A.L.Vivan,
L.M.Gava,
R.Dias,
R.G.Ducati,
L.A.Basso,
D.S.Santos,
W.F.de Azevedo.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Human purine nucleoside phosphorylase (HsPNP) is a target for inhibitor
development aiming at T-cell immune response modulation. In this work, we report
the development of a new set of empirical scoring functions and its application
to evaluate binding affinities and docking results. To test these new functions,
we solved the structure of HsPNP and 2-mercapto-4(3H)-quinazolinone (HsPNP:MQU)
binary complex at 2.7A resolution using synchrotron radiation, and used these
functions to predict ligand position obtained in docking simulations. We also
employed molecular dynamics simulations to analyze HsPNP in two conditions, as
apoenzyme and in the binary complex form, in order to assess the structural
features responsible for stability. Analysis of the structural differences
between systems provides explanation for inhibitor binding. The use of these
scoring functions to evaluate binding affinities and molecular docking results
may be used to guide future efforts on virtual screening focused on HsPNP.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
F.B.Zanchi,
R.A.Caceres,
R.G.Stabeli,
and
W.F.de Azevedo
(2010).
Molecular dynamics studies of a hexameric purine nucleoside phosphorylase.
|
| |
J Mol Model,
16,
543-550.
|
 |
|
|
|
|
 |
M.L.Bellows,
and
C.A.Floudas
(2010).
Computational methods for de novo protein design and its applications to the human immunodeficiency virus 1, purine nucleoside phosphorylase, ubiquitin specific protease 7, and histone demethylases.
|
| |
Curr Drug Targets,
11,
264-278.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

