 |
PDBsum entry 3css

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.1.31
- 6-phosphogluconolactonase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pentose Phosphate Pathway (early stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
6-phospho-D-glucono-1,5-lactone + H2O = 6-phospho-D-gluconate + H+
|
 |
 |
 |
 |
 |
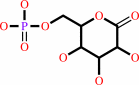 6-phospho-D-glucono-1,5-lactone
6-phospho-D-glucono-1,5-lactone
|
+
|
H2O
|
=
|
 6-phospho-D-gluconate
6-phospho-D-gluconate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
62:439-450
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Optimal description of a protein structure in terms of multiple groups undergoing TLS motion.
|
|
J.Painter,
E.A.Merritt.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
A single protein crystal structure contains information about dynamic properties
of the protein as well as providing a static view of one three-dimensional
conformation. This additional information is to be found in the distribution of
observed electron density about the mean position of each atom. It is general
practice to account for this by refining a separate atomic displacement
parameter (ADP) for each atomic center. However, these same displacements are
often described well by simpler models based on TLS
(translation/libration/screw) rigid-body motion of large groups of atoms, for
example interdomain hinge motion. A procedure, TLSMD, has been developed that
analyzes the distribution of ADPs in a previously refined protein crystal
structure in order to generate optimal multi-group TLS descriptions of the
constituent protein chains. TLSMD is applicable to crystal structures at any
resolution. The models generated by TLSMD analysis can significantly improve the
standard crystallographic residuals R and R(free) and can reveal intrinsic
dynamic properties of the protein.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 The graph constructed in stage 2 of the TLSMD
algorithm for finding the optimal segmentation of a peptide
chain into multiple contiguous TLS groups.
|
 |
Figure 4.
Figure 4 Crystallographic refinement of multi-group TLS models
for L. mexicana initiation factor 5A. This 2.7 Å data set
was collected from a SeMet form of the protein. The R and
R[free] residuals from conventional refinement with individual
isotopic B factors are shown as thin horizontal lines. The
dotted lines track the residuals from pure TLS refinement
(individual atoms are described only the TLS model, with no
individual B[iso] component). The heavy solid lines correspond
to refinement of TLS models supplemented by individual B[iso]
components.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2006,
62,
439-450)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.L.Morgan,
J.Strumillo,
and
J.Zimmer
(2013).
Crystallographic snapshot of cellulose synthesis and membrane translocation.
|
| |
Nature,
493,
181-186.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Alon,
I.Grossman,
Y.Gat,
V.K.Kodali,
F.DiMaio,
T.Mehlman,
G.Haran,
D.Baker,
C.Thorpe,
and
D.Fass
(2012).
The dynamic disulphide relay of quiescin sulphydryl oxidase.
|
| |
Nature,
488,
414-418.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Krishnamurthy,
and
E.Gouaux
(2012).
X-ray structures of LeuT in substrate-free outward-open and apo inward-open states.
|
| |
Nature,
481,
469-474.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Payandeh,
T.M.Gamal El-Din,
T.Scheuer,
N.Zheng,
and
W.A.Catterall
(2012).
Crystal structure of a voltage-gated sodium channel in two potentially inactivated states.
|
| |
Nature,
486,
135-139.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.A.Yates,
S.Fleurdépine,
O.S.Rissland,
L.De Colibus,
K.Harlos,
C.J.Norbury,
and
R.J.Gilbert
(2012).
Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase.
|
| |
Nat Struct Mol Biol,
19,
782-787.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.M.Bandaranayake,
D.Ungureanu,
Y.Shan,
D.E.Shaw,
O.Silvennoinen,
and
S.R.Hubbard
(2012).
Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F.
|
| |
Nat Struct Mol Biol,
19,
754-759.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.C.Chao,
K.Kulkarni,
Z.Zhang,
E.H.Kong,
and
D.Barford
(2012).
Structure of the mitotic checkpoint complex.
|
| |
Nature,
484,
208-213.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.M.Lunde,
M.Hörner,
and
A.Meinhart
(2011).
Structural insights into cis element recognition of non-polyadenylated RNAs by the Nab3-RRM.
|
| |
Nucleic Acids Res,
39,
337-346.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Xu,
J.C.Pizarro,
M.A.Holmes,
C.McBeth,
V.Groh,
T.Spies,
and
R.K.Strong
(2011).
Crystal structure of a gammadelta T-cell receptor specific for the human MHC class I homolog MICA.
|
| |
Proc Natl Acad Sci U S A,
108,
2414-2419.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.An,
S.Lovell,
M.R.Kanost,
K.P.Battaile,
and
K.Michel
(2011).
Crystal structure of native Anopheles gambiae serpin-2, a negative regulator of melanization in mosquitoes.
|
| |
Proteins,
79,
1999-2003.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Close,
S.J.Johnson,
M.A.Sdano,
S.M.McDonald,
H.Robinson,
T.Formosa,
and
C.P.Hill
(2011).
Crystal structures of the S. cerevisiae Spt6 core and C-terminal tandem SH2 domain.
|
| |
J Mol Biol,
408,
697-713.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.Nachon,
E.Carletti,
M.Wandhammer,
Y.Nicolet,
L.M.Schopfer,
P.Masson,
and
O.Lockridge
(2011).
X-ray crystallographic snapshots of reaction intermediates in the G117H mutant of human butyrylcholinesterase, a nerve agent target engineered into a catalytic bioscavenger.
|
| |
Biochem J,
434,
73-82.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.M.Alushin,
D.Jane,
and
M.L.Mayer
(2011).
Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
|
| |
Neuropharmacology,
60,
126-134.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.N.Murshudov,
P.Skubák,
A.A.Lebedev,
N.S.Pannu,
R.A.Steiner,
R.A.Nicholls,
M.D.Winn,
F.Long,
and
A.A.Vagin
(2011).
REFMAC5 for the refinement of macromolecular crystal structures.
|
| |
Acta Crystallogr D Biol Crystallogr,
67,
355-367.
|
 |
|
|
|
|
 |
G.T.Lountos,
J.E.Tropea,
and
D.S.Waugh
(2011).
Structure of human dual-specificity phosphatase 27 at 2.38 Å resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
67,
471-479.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.N.Kirouac,
and
H.Ling
(2011).
Unique active site promotes error-free replication opposite an 8-oxo-guanine lesion by human DNA polymerase iota.
|
| |
Proc Natl Acad Sci U S A,
108,
3210-3215.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.B.Lazarus,
Y.Nam,
J.Jiang,
P.Sliz,
and
S.Walker
(2011).
Structure of human O-GlcNAc transferase and its complex with a peptide substrate.
|
| |
Nature,
469,
564-567.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Yang,
W.Ge,
R.Chowdhury,
T.D.Claridge,
H.B.Kramer,
B.Schmierer,
M.A.McDonough,
L.Gong,
B.M.Kessler,
P.J.Ratcliffe,
M.L.Coleman,
and
C.J.Schofield
(2011).
Asparagine and Aspartate Hydroxylation of the Cytoskeletal Ankyrin Family Is Catalyzed by Factor-inhibiting Hypoxia-inducible Factor.
|
| |
J Biol Chem,
286,
7648-7660.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Empadinhas,
P.J.Pereira,
L.Albuquerque,
J.Costa,
B.Sá-Moura,
A.T.Marques,
S.Macedo-Ribeiro,
and
M.S.da Costa
(2011).
Functional and structural characterization of a novel mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus reveals its dual substrate specificity.
|
| |
Mol Microbiol,
79,
76-93.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.Fedorov,
K.Huber,
A.Eisenreich,
P.Filippakopoulos,
O.King,
A.N.Bullock,
D.Szklarczyk,
L.J.Jensen,
D.Fabbro,
J.Trappe,
U.Rauch,
F.Bracher,
and
S.Knapp
(2011).
Specific CLK inhibitors from a novel chemotype for regulation of alternative splicing.
|
| |
Chem Biol,
18,
67-76.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Jauch,
S.H.Choo,
C.K.Ng,
and
P.R.Kolatkar
(2011).
Crystal structure of the dimeric Oct6 (POU3f1) POU domain bound to palindromic MORE DNA.
|
| |
Proteins,
79,
674-677.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.R.Thomas,
C.A.Keller,
A.Szyk,
J.R.Cannon,
and
N.A.Laronde-Leblanc
(2011).
Structural insight into the functional mechanism of Nep1/Emg1 N1-specific pseudouridine methyltransferase in ribosome biogenesis.
|
| |
Nucleic Acids Res,
39,
2445-2457.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.S.Rizk,
M.Paduch,
J.H.Heithaus,
E.M.Duguid,
A.Sandstrom,
and
A.A.Kossiakoff
(2011).
Allosteric control of ligand-binding affinity using engineered conformation-specific effector proteins.
|
| |
Nat Struct Mol Biol,
18,
437-442.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Welti,
S.Kühn,
I.D'Angelo,
B.Brügger,
D.Kaufmann,
and
K.Scheffzek
(2011).
Structural and biochemical consequences of NF1 associated nontruncating mutations in the Sec14-PH module of neurofibromin.
|
| |
Hum Mutat,
32,
191-197.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Ostendorp,
J.Diez,
C.W.Heizmann,
and
G.Fritz
(2011).
The crystal structures of human S100B in the zinc- and calcium-loaded state at three pH values reveal zinc ligand swapping.
|
| |
Biochim Biophys Acta,
1813,
1083-1091.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.W.James,
N.Frias-Staheli,
J.P.Bacik,
J.M.Levingston Macleod,
M.Khajehpour,
A.García-Sastre,
and
B.L.Mark
(2011).
Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease.
|
| |
Proc Natl Acad Sci U S A,
108,
2222-2227.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.W.Cheng,
J.G.McCoy,
A.N.Thompson,
C.G.Nichols,
and
C.M.Nimigean
(2011).
Mechanism for selectivity-inactivation coupling in KcsA potassium channels.
|
| |
Proc Natl Acad Sci U S A,
108,
5272-5277.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.J.Schoeffler,
A.P.May,
and
J.M.Berger
(2010).
A domain insertion in Escherichia coli GyrB adopts a novel fold that plays a critical role in gyrase function.
|
| |
Nucleic Acids Res,
38,
7830-7844.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.P.Yeh,
P.Abdubek,
T.Astakhova,
H.L.Axelrod,
C.Bakolitsa,
X.Cai,
D.Carlton,
C.Chen,
H.J.Chiu,
M.Chiu,
T.Clayton,
D.Das,
M.C.Deller,
L.Duan,
K.Ellrott,
C.L.Farr,
J.Feuerhelm,
J.C.Grant,
A.Grzechnik,
G.W.Han,
L.Jaroszewski,
K.K.Jin,
H.E.Klock,
M.W.Knuth,
P.Kozbial,
S.S.Krishna,
A.Kumar,
W.W.Lam,
D.Marciano,
D.McMullan,
M.D.Miller,
A.T.Morse,
E.Nigoghossian,
A.Nopakun,
L.Okach,
C.Puckett,
R.Reyes,
H.J.Tien,
C.B.Trame,
H.van den Bedem,
D.Weekes,
T.Wooten,
Q.Xu,
K.O.Hodgson,
J.Wooley,
M.A.Elsliger,
A.M.Deacon,
A.Godzik,
S.A.Lesley,
and
I.A.Wilson
(2010).
Structure of Bacteroides thetaiotaomicron BT2081 at 2.05 Å resolution: the first structural representative of a new protein family that may play a role in carbohydrate metabolism.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
66,
1287-1296.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.E.Correia,
Y.E.Ban,
M.A.Holmes,
H.Xu,
K.Ellingson,
Z.Kraft,
C.Carrico,
E.Boni,
D.N.Sather,
C.Zenobia,
K.Y.Burke,
T.Bradley-Hewitt,
J.F.Bruhn-Johannsen,
O.Kalyuzhniy,
D.Baker,
R.K.Strong,
L.Stamatatos,
and
W.R.Schief
(2010).
Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope.
|
| |
Structure,
18,
1116-1126.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Ford,
A.T.Rêgo,
T.J.Ragan,
J.Pinkner,
K.Dodson,
P.C.Driscoll,
S.Hultgren,
and
G.Waksman
(2010).
Structural homology between the C-terminal domain of the PapC usher and its plug.
|
| |
J Bacteriol,
192,
1824-1831.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Bakolitsa,
A.Kumar,
D.McMullan,
S.S.Krishna,
M.D.Miller,
D.Carlton,
R.Najmanovich,
P.Abdubek,
T.Astakhova,
H.J.Chiu,
T.Clayton,
M.C.Deller,
L.Duan,
Y.Elias,
J.Feuerhelm,
J.C.Grant,
S.K.Grzechnik,
G.W.Han,
L.Jaroszewski,
K.K.Jin,
H.E.Klock,
M.W.Knuth,
P.Kozbial,
D.Marciano,
A.T.Morse,
E.Nigoghossian,
L.Okach,
S.Oommachen,
J.Paulsen,
R.Reyes,
C.L.Rife,
C.V.Trout,
H.van den Bedem,
D.Weekes,
A.White,
Q.Xu,
K.O.Hodgson,
J.Wooley,
M.A.Elsliger,
A.M.Deacon,
A.Godzik,
S.A.Lesley,
and
I.A.Wilson
(2010).
The structure of the first representative of Pfam family PF06475 reveals a new fold with possible involvement in glycolipid metabolism.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
66,
1211-1217.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.L.Ng,
D.G.Waterman,
A.A.Antson,
and
M.Ortiz-Lombardía
(2010).
Structure of the Methanothermobacter thermautotrophicus exosome RNase PH ring.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
522-528.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.M.Bianchetti,
G.C.Blouin,
E.Bitto,
J.S.Olson,
and
G.N.Phillips
(2010).
The structure and NO binding properties of the nitrophorin-like heme-binding protein from Arabidopsis thaliana gene locus At1g79260.1.
|
| |
Proteins,
78,
917-931.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.R.Bourne,
E.W.Barrow,
R.A.Bunce,
P.C.Bourne,
K.D.Berlin,
and
W.W.Barrow
(2010).
Inhibition of antibiotic-resistant Staphylococcus aureus by the broad-spectrum dihydrofolate reductase inhibitor RAB1.
|
| |
Antimicrob Agents Chemother,
54,
3825-3833.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.R.Kimberlin,
Z.A.Bornholdt,
S.Li,
V.L.Woods,
I.J.MacRae,
and
E.O.Saphire
(2010).
Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
|
| |
Proc Natl Acad Sci U S A,
107,
314-319.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.S.Anthony,
H.R.Corradi,
S.L.Schwager,
P.Redelinghuys,
D.Georgiadis,
V.Dive,
K.R.Acharya,
and
E.D.Sturrock
(2010).
The N domain of human angiotensin-I-converting enzyme: the role of N-glycosylation and the crystal structure in complex with an N domain-specific phosphinic inhibitor, RXP407.
|
| |
J Biol Chem,
285,
35685-35693.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.W.Vander Kooi,
L.Ren,
P.Xu,
M.D.Ohi,
K.L.Gould,
and
W.J.Chazin
(2010).
The Prp19 WD40 domain contains a conserved protein interaction region essential for its function.
|
| |
Structure,
18,
584-593.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Koutsioulis,
A.Lyskowski,
S.Mäki,
E.Guthrie,
G.Feller,
V.Bouriotis,
and
P.Heikinheimo
(2010).
Coordination sphere of the third metal site is essential to the activity and metal selectivity of alkaline phosphatases.
|
| |
Protein Sci,
19,
75-84.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.W.Leung,
K.C.Prins,
D.M.Borek,
M.Farahbakhsh,
J.M.Tufariello,
P.Ramanan,
J.C.Nix,
L.A.Helgeson,
Z.Otwinowski,
R.B.Honzatko,
C.F.Basler,
and
G.K.Amarasinghe
(2010).
Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
|
| |
Nat Struct Mol Biol,
17,
165-172.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.T.Larson,
D.G.Mudeppa,
J.R.Gillespie,
N.Mueller,
A.J.Napuli,
J.A.Arif,
J.Ross,
T.L.Arakaki,
A.Lauricella,
G.Detitta,
J.Luft,
F.Zucker,
C.L.Verlinde,
E.Fan,
W.C.Van Voorhis,
F.S.Buckner,
P.K.Rathod,
W.G.Hol,
and
E.A.Merritt
(2010).
The crystal structure and activity of a putative trypanosomal nucleoside phosphorylase reveal it to be a homodimeric uridine phosphorylase.
|
| |
J Mol Biol,
396,
1244-1259.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.C.Peterson,
E.S.Burgie,
S.Y.Park,
D.R.Jensen,
J.J.Weiner,
C.A.Bingman,
C.E.Chang,
S.R.Cutler,
G.N.Phillips,
and
B.F.Volkman
(2010).
Structural basis for selective activation of ABA receptors.
|
| |
Nat Struct Mol Biol,
17,
1109-1113.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.Zucker,
P.C.Champ,
and
E.A.Merritt
(2010).
Validation of crystallographic models containing TLS or other descriptions of anisotropy.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
889-900.
|
 |
|
|
|
|
 |
G.Askarieh,
M.Hedhammar,
K.Nordling,
A.Saenz,
C.Casals,
A.Rising,
J.Johansson,
and
S.D.Knight
(2010).
Self-assembly of spider silk proteins is controlled by a pH-sensitive relay.
|
| |
Nature,
465,
236-238.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Manina,
M.Bellinzoni,
M.R.Pasca,
J.Neres,
A.Milano,
A.L.Ribeiro,
S.Buroni,
H.Skovierová,
P.Dianišková,
K.Mikušová,
J.Marák,
V.Makarov,
D.Giganti,
A.Haouz,
A.P.Lucarelli,
G.Degiacomi,
A.Piazza,
L.R.Chiarelli,
E.De Rossi,
E.Salina,
S.T.Cole,
P.M.Alzari,
and
G.Riccardi
(2010).
Biological and structural characterization of the Mycobacterium smegmatis nitroreductase NfnB, and its role in benzothiazinone resistance.
|
| |
Mol Microbiol,
77,
1172-1185.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.L.Schubert,
Q.Zhai,
V.Sandrin,
D.M.Eckert,
M.Garcia-Maya,
L.Saul,
W.I.Sundquist,
R.A.Steiner,
and
C.P.Hill
(2010).
Structural and functional studies on the extracellular domain of BST2/tetherin in reduced and oxidized conformations.
|
| |
Proc Natl Acad Sci U S A,
107,
17951-17956.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Xu,
L.Song,
M.Kim,
M.A.Holmes,
Z.Kraft,
G.Sellhorn,
E.L.Reinherz,
L.Stamatatos,
and
R.K.Strong
(2010).
Interactions between lipids and human anti-HIV antibody 4E10 can be reduced without ablating neutralizing activity.
|
| |
J Virol,
84,
1076-1088.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Ashworth,
G.K.Taylor,
J.J.Havranek,
S.A.Quadri,
B.L.Stoddard,
and
D.Baker
(2010).
Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs.
|
| |
Nucleic Acids Res,
38,
5601-5608.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.B.Bruning,
A.A.Parent,
G.Gil,
M.Zhao,
J.Nowak,
M.C.Pace,
C.L.Smith,
P.V.Afonine,
P.D.Adams,
J.A.Katzenellenbogen,
and
K.W.Nettles
(2010).
Coupling of receptor conformation and ligand orientation determine graded activity.
|
| |
Nat Chem Biol,
6,
837-843.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.C.Grigg,
J.Cheung,
D.E.Heinrichs,
and
M.E.Murphy
(2010).
Specificity of Staphyloferrin B recognition by the SirA receptor from Staphylococcus aureus.
|
| |
J Biol Chem,
285,
34579-34588.
|
 |
|
|
|
|
 |
J.Hafner,
and
W.Zheng
(2010).
Optimal modeling of atomic fluctuations in protein crystal structures for weak crystal contact interactions.
|
| |
J Chem Phys,
132,
014111.
|
 |
|
|
|
|
 |
J.K.Forwood,
A.Lange,
U.Zachariae,
M.Marfori,
C.Preast,
H.Grubmüller,
M.Stewart,
A.H.Corbett,
and
B.Kobe
(2010).
Quantitative structural analysis of importin-β flexibility: paradigm for solenoid protein structures.
|
| |
Structure,
18,
1171-1183.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Lowther,
B.A.Yard,
K.A.Johnson,
L.G.Carter,
V.T.Bhat,
M.C.Raman,
D.J.Clarke,
B.Ramakers,
S.A.McMahon,
J.H.Naismith,
and
D.J.Campopiano
(2010).
Inhibition of the PLP-dependent enzyme serine palmitoyltransferase by cycloserine: evidence for a novel decarboxylative mechanism of inactivation.
|
| |
Mol Biosyst,
6,
1682-1693.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.C.Prins,
S.Delpeut,
D.W.Leung,
O.Reynard,
V.A.Volchkova,
S.P.Reid,
P.Ramanan,
W.B.Cárdenas,
G.K.Amarasinghe,
V.E.Volchkov,
and
C.F.Basler
(2010).
Mutations abrogating VP35 interaction with double-stranded RNA render Ebola virus avirulent in guinea pigs.
|
| |
J Virol,
84,
3004-3015.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Sisinni,
L.Cendron,
G.Favaro,
and
G.Zanotti
(2010).
Helicobacter pylori acidic stress response factor HP1286 is a YceI homolog with new binding specificity.
|
| |
FEBS J,
277,
1896-1905.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Yu,
Y.Wang,
S.Huang,
J.Wang,
Z.Deng,
Q.Zhang,
W.Wu,
X.Zhang,
Z.Liu,
W.Gong,
and
Z.Chen
(2010).
Structural insights into a novel histone demethylase PHF8.
|
| |
Cell Res,
20,
166-173.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.A.Arbing,
M.Kaufmann,
T.Phan,
S.Chan,
D.Cascio,
and
D.Eisenberg
(2010).
The crystal structure of the Mycobacterium tuberculosis Rv3019c-Rv3020c ESX complex reveals a domain-swapped heterotetramer.
|
| |
Protein Sci,
19,
1692-1703.
|
 |
|
|
|
|
 |
M.Haffke,
A.Menzel,
Y.Carius,
D.Jahn,
and
D.W.Heinz
(2010).
Structures of the nucleotide-binding domain of the human ABCB6 transporter and its complexes with nucleotides.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
979-987.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Resch,
E.Schiltz,
F.Titgemeyer,
and
Y.A.Muller
(2010).
Insight into the induction mechanism of the GntR/HutC bacterial transcription regulator YvoA.
|
| |
Nucleic Acids Res,
38,
2485-2497.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
O.Spadiut,
T.C.Tan,
I.Pisanelli,
D.Haltrich,
and
C.Divne
(2010).
Importance of the gating segment in the substrate-recognition loop of pyranose 2-oxidase.
|
| |
FEBS J,
277,
2892-2909.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Filippakopoulos,
A.Low,
T.D.Sharpe,
J.Uppenberg,
S.Yao,
Z.Kuang,
P.Savitsky,
R.S.Lewis,
S.E.Nicholson,
R.S.Norton,
and
A.N.Bullock
(2010).
Structural basis for Par-4 recognition by the SPRY domain- and SOCS box-containing proteins SPSB1, SPSB2, and SPSB4.
|
| |
J Mol Biol,
401,
389-402.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.J.Holland,
and
T.Hollis
(2010).
Structural and mutational analysis of Escherichia coli AlkB provides insight into substrate specificity and DNA damage searching.
|
| |
PLoS One,
5,
e8680.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Leone,
D.Comoletti,
G.Ferracci,
S.Conrod,
S.U.Garcia,
P.Taylor,
Y.Bourne,
and
P.Marchot
(2010).
Structural insights into the exquisite selectivity of neurexin/neuroligin synaptic interactions.
|
| |
EMBO J,
29,
2461-2471.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.R.Elliott,
B.T.Goult,
P.M.Kopp,
N.Bate,
J.G.Grossmann,
G.C.Roberts,
D.R.Critchley,
and
I.L.Barsukov
(2010).
The Structure of the talin head reveals a novel extended conformation of the FERM domain.
|
| |
Structure,
18,
1289-1299.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Diskin,
P.M.Marcovecchio,
and
P.J.Bjorkman
(2010).
Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity.
|
| |
Nat Struct Mol Biol,
17,
608-613.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.M.Kennan,
W.Wong,
O.P.Dhungyel,
X.Han,
D.Wong,
D.Parker,
C.J.Rosado,
R.H.Law,
S.McGowan,
S.B.Reeve,
V.Levina,
G.A.Powers,
R.N.Pike,
S.P.Bottomley,
A.I.Smith,
I.Marsh,
R.J.Whittington,
J.C.Whisstock,
C.J.Porter,
and
J.I.Rood
(2010).
The subtilisin-like protease AprV2 is required for virulence and uses a novel disulphide-tethered exosite to bind substrates.
|
| |
PLoS Pathog,
6,
e1001210.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.B.Hari,
C.Byeon,
J.J.Lavinder,
and
T.J.Magliery
(2010).
Cysteine-free Rop: a four-helix bundle core mutant has wild-type stability and structure but dramatically different unfolding kinetics.
|
| |
Protein Sci,
19,
670-679.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.D'Arcy,
O.R.Davies,
T.L.Blundell,
and
V.M.Bolanos-Garcia
(2010).
Defining the molecular basis of BubR1 kinetochore interactions and APC/C-CDC20 inhibition.
|
| |
J Biol Chem,
285,
14764-14776.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.K.Menon,
B.J.Eilers,
M.J.Young,
and
C.M.Lawrence
(2010).
The crystal structure of D212 from sulfolobus spindle-shaped virus ragged hills reveals a new member of the PD-(D/E)XK nuclease superfamily.
|
| |
J Virol,
84,
5890-5897.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Müller,
G.Zocher,
A.Steinle,
and
T.Stehle
(2010).
Structure of the HCMV UL16-MICB complex elucidates select binding of a viral immunoevasin to diverse NKG2D ligands.
|
| |
PLoS Pathog,
6,
e1000723.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.M.McDonald,
D.Close,
H.Xin,
T.Formosa,
and
C.P.Hill
(2010).
Structure and biological importance of the Spn1-Spt6 interaction, and its regulatory role in nucleosome binding.
|
| |
Mol Cell,
40,
725-735.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.M.Shandilya,
M.N.Nalam,
E.A.Nalivaika,
P.J.Gross,
J.C.Valesano,
K.Shindo,
M.Li,
M.Munson,
W.E.Royer,
E.Harjes,
T.Kono,
H.Matsuo,
R.S.Harris,
M.Somasundaran,
and
C.A.Schiffer
(2010).
Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces.
|
| |
Structure,
18,
28-38.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Kowatz,
J.P.Morrison,
M.E.Tanner,
and
J.H.Naismith
(2010).
The crystal structure of the Y140F mutant of ADP-L-glycero-D-manno-heptose 6-epimerase bound to ADP-beta-D-mannose suggests a one base mechanism.
|
| |
Protein Sci,
19,
1337-1343.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Z.Chen,
D.Borek,
S.B.Padrick,
T.S.Gomez,
Z.Metlagel,
A.M.Ismail,
J.Umetani,
D.D.Billadeau,
Z.Otwinowski,
and
M.K.Rosen
(2010).
Structure and control of the actin regulatory WAVE complex.
|
| |
Nature,
468,
533-538.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Chaikuad,
and
R.L.Brady
(2009).
Conservation of structure and activity in Plasmodium purine nucleoside phosphorylases.
|
| |
BMC Struct Biol,
9,
42.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Goulet,
S.Spinelli,
S.Blangy,
H.van Tilbeurgh,
N.Leulliot,
T.Basta,
D.Prangishvili,
C.Cambillau,
and
V.Campanacci
(2009).
The thermo- and acido-stable ORF-99 from the archaeal virus AFV1.
|
| |
Protein Sci,
18,
1316-1320.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.J.Kimple,
M.Soundararajan,
S.Q.Hutsell,
A.K.Roos,
D.J.Urban,
V.Setola,
B.R.Temple,
B.L.Roth,
S.Knapp,
F.S.Willard,
and
D.P.Siderovski
(2009).
Structural Determinants of G-protein {alpha} Subunit Selectivity by Regulator of G-protein Signaling 2 (RGS2).
|
| |
J Biol Chem,
284,
19402-19411.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.K.Kaiser,
M.C.Clifton,
B.W.Shen,
and
B.L.Stoddard
(2009).
The structure of a bacterial DUF199/WhiA protein: domestication of an invasive endonuclease.
|
| |
Structure,
17,
1368-1376.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Bebrone,
H.Delbrück,
M.B.Kupper,
P.Schlömer,
C.Willmann,
J.M.Frère,
R.Fischer,
M.Galleni,
and
K.M.Hoffmann
(2009).
The structure of the dizinc subclass B2 metallo-beta-lactamase CphA reveals that the second inhibitory zinc ion binds in the histidine site.
|
| |
Antimicrob Agents Chemother,
53,
4464-4471.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Han,
K.Kim,
Y.Kim,
Y.Kang,
J.Y.Lee,
and
Y.Kim
(2009).
Crystal structure of the N-terminal domain of anaphase-promoting complex subunit 7.
|
| |
J Biol Chem,
284,
15137-15146.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Veesler,
B.Dreier,
S.Blangy,
J.Lichière,
D.Tremblay,
S.Moineau,
S.Spinelli,
M.Tegoni,
A.Plückthun,
V.Campanacci,
and
C.Cambillau
(2009).
Crystal structure and function of a DARPin neutralizing inhibitor of lactococcal phage TP901-1: comparison of DARPin and camelid VHH binding mode.
|
| |
J Biol Chem,
284,
30718-30726.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.M.Warren,
H.Huang,
E.Fanning,
W.J.Chazin,
and
B.F.Eichman
(2009).
Physical interactions between Mcm10, DNA, and DNA polymerase alpha.
|
| |
J Biol Chem,
284,
24662-24672.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Sauvage,
A.Zervosen,
G.Dive,
R.Herman,
A.Amoroso,
B.Joris,
E.Fonzé,
R.F.Pratt,
A.Luxen,
P.Charlier,
and
F.Kerff
(2009).
Structural basis of the inhibition of class A beta-lactamases and penicillin-binding proteins by 6-beta-iodopenicillanate.
|
| |
J Am Chem Soc,
131,
15262-15269.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.T.Larson,
F.Parussini,
M.H.Huynh,
J.D.Giebel,
A.M.Kelley,
L.Zhang,
M.Bogyo,
E.A.Merritt,
and
V.B.Carruthers
(2009).
Toxoplasma gondii cathepsin L is the primary target of the invasion-inhibitory compound morpholinurea-leucyl-homophenyl-vinyl sulfone phenyl.
|
| |
J Biol Chem,
284,
26839-26850.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.B.Rha,
G.Wu,
S.E.Shoelson,
and
Y.I.Chi
(2009).
Multiple binding modes between HNF4alpha and the LXXLL motifs of PGC-1alpha lead to full activation.
|
| |
J Biol Chem,
284,
35165-35176.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.J.Lee,
B.Rakić,
M.Gilbert,
W.W.Wakarchuk,
S.G.Withers,
and
N.C.Strynadka
(2009).
Structural and kinetic characterizations of the polysialic acid O-acetyltransferase OatWY from Neisseria meningitidis.
|
| |
J Biol Chem,
284,
24501-24511.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.von Moeller,
C.Basquin,
and
E.Conti
(2009).
The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner.
|
| |
Nat Struct Mol Biol,
16,
247-254.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Westwood,
D.M.Cheary,
J.E.Baxter,
M.W.Richards,
R.L.van Montfort,
A.M.Fry,
and
R.Bayliss
(2009).
Insights into the conformational variability and regulation of human Nek2 kinase.
|
| |
J Mol Biol,
386,
476-485.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.A.Garnett,
Y.Liu,
E.Leon,
S.A.Allman,
N.Friedrich,
S.Saouros,
S.Curry,
D.Soldati-Favre,
B.G.Davis,
T.Feizi,
and
S.Matthews
(2009).
Detailed insights from microarray and crystallographic studies into carbohydrate recognition by microneme protein 1 (MIC1) of Toxoplasma gondii.
|
| |
Protein Sci,
18,
1935-1947.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Abendroth,
D.D.Mitchell,
K.V.Korotkov,
T.L.Johnson,
A.Kreger,
M.Sandkvist,
and
W.G.Hol
(2009).
The three-dimensional structure of the cytoplasmic domains of EpsF from the type 2 secretion system of Vibrio cholerae.
|
| |
J Struct Biol,
166,
303-315.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.D.Ho,
R.Yeh,
A.Sandstrom,
I.Chorny,
W.E.Harries,
R.A.Robbins,
L.J.Miercke,
and
R.M.Stroud
(2009).
Crystal structure of human aquaporin 4 at 1.8 A and its mechanism of conductance.
|
| |
Proc Natl Acad Sci U S A,
106,
7437-7442.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.G.McCoy,
H.D.Johnson,
S.Singh,
C.A.Bingman,
I.K.Lei,
J.S.Thorson,
and
G.N.Phillips
(2009).
Structural characterization of CalO2: a putative orsellinic acid P450 oxidase in the calicheamicin biosynthetic pathway.
|
| |
Proteins,
74,
50-60.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Guhaniyogi,
I.Sohar,
K.Das,
A.M.Stock,
and
P.Lobel
(2009).
Crystal Structure and Autoactivation Pathway of the Precursor Form of Human Tripeptidyl-peptidase 1, the Enzyme Deficient in Late Infantile Ceroid Lipofuscinosis.
|
| |
J Biol Chem,
284,
3985-3997.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.K.Archbold,
W.A.Macdonald,
S.Gras,
L.K.Ely,
J.J.Miles,
M.J.Bell,
R.M.Brennan,
T.Beddoe,
M.C.Wilce,
C.S.Clements,
A.W.Purcell,
J.McCluskey,
S.R.Burrows,
and
J.Rossjohn
(2009).
Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition.
|
| |
J Exp Med,
206,
209-219.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Kumar,
P.Schuck,
R.Jin,
and
M.L.Mayer
(2009).
The N-terminal domain of GluR6-subtype glutamate receptor ion channels.
|
| |
Nat Struct Mol Biol,
16,
631-638.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.V.Korotkov,
E.Pardon,
J.Steyaert,
and
W.G.Hol
(2009).
Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody.
|
| |
Structure,
17,
255-265.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.V.Korotkov,
M.D.Gray,
A.Kreger,
S.Turley,
M.Sandkvist,
and
W.G.Hol
(2009).
Calcium is essential for the major pseudopilin in the type 2 secretion system.
|
| |
J Biol Chem,
284,
25466-25470.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.I.Robins,
A.H.Williams,
and
C.R.Raetz
(2009).
Structural basis for the sugar nucleotide and acyl-chain selectivity of Leptospira interrogans LpxA.
|
| |
Biochemistry,
48,
6191-6201.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Min,
Z.Jin,
L.Caldovic,
H.Morizono,
N.M.Allewell,
M.Tuchman,
and
D.Shi
(2009).
Mechanism of Allosteric Inhibition of N-Acetyl-L-glutamate Synthase by L-Arginine.
|
| |
J Biol Chem,
284,
4873-4880.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.E.Jackrel,
R.Valverde,
and
L.Regan
(2009).
Redesign of a protein-peptide interaction: characterization and applications.
|
| |
Protein Sci,
18,
762-774.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.J.Maher,
S.Akimoto,
M.Iwata,
K.Nagata,
Y.Hori,
M.Yoshida,
S.Yokoyama,
S.Iwata,
and
K.Yokoyama
(2009).
Crystal structure of A3B3 complex of V-ATPase from Thermus thermophilus.
|
| |
EMBO J,
28,
3771-3779.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.S.Hanes,
K.M.Jude,
J.M.Berger,
R.A.Bonomo,
and
T.M.Handel
(2009).
Structural and biochemical characterization of the interaction between KPC-2 beta-lactamase and beta-lactamase inhibitor protein.
|
| |
Biochemistry,
48,
9185-9193.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.T.Sung,
Y.T.Lai,
C.Y.Huang,
L.Y.Chou,
H.W.Shih,
W.C.Cheng,
C.H.Wong,
and
C.Ma
(2009).
Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli.
|
| |
Proc Natl Acad Sci U S A,
106,
8824-8829.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.W.Richards,
L.O'Regan,
C.Mas-Droux,
J.M.Blot,
J.Cheung,
S.Hoelder,
A.M.Fry,
and
R.Bayliss
(2009).
An autoinhibitory tyrosine motif in the cell-cycle-regulated Nek7 kinase is released through binding of Nek9.
|
| |
Mol Cell,
36,
560-570.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Barison,
L.Cendron,
A.Trento,
A.Angelini,
and
G.Zanotti
(2009).
Structural and mutational analysis of TenA protein (HP1287) from the Helicobacter pylori thiamin salvage pathway - evidence of a different substrate specificity.
|
| |
FEBS J,
276,
6227-6235.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
O.C.Ezezika,
N.S.Younger,
J.Lu,
D.A.Kaiser,
Z.A.Corbin,
B.J.Nolen,
D.R.Kovar,
and
T.D.Pollard
(2009).
Incompatibility with Formin Cdc12p Prevents Human Profilin from Substituting for Fission Yeast Profilin: INSIGHTS FROM CRYSTAL STRUCTURES OF FISSION YEAST PROFILIN.
|
| |
J Biol Chem,
284,
2088-2097.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.Spadiut,
C.Leitner,
C.Salaheddin,
B.Varga,
B.G.Vertessy,
T.C.Tan,
C.Divne,
and
D.Haltrich
(2009).
Improving thermostability and catalytic activity of pyranose 2-oxidase from Trametes multicolor by rational and semi-rational design.
|
| |
FEBS J,
276,
776-792.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.Spadiut,
K.Radakovits,
I.Pisanelli,
C.Salaheddin,
M.Yamabhai,
T.C.Tan,
C.Divne,
and
D.Haltrich
(2009).
A thermostable triple mutant of pyranose 2-oxidase from Trametes multicolor with improved properties for biotechnological applications.
|
| |
Biotechnol J,
4,
525-534.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.A.Meyer,
P.Ye,
M.H.Suh,
M.Zhang,
and
J.Fu
(2009).
Structure of the 12-Subunit RNA Polymerase II Refined with the Aid of Anomalous Diffraction Data.
|
| |
J Biol Chem,
284,
12933-12939.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Aranda,
H.Cai,
C.E.Worley,
E.J.Levin,
R.Li,
J.S.Olson,
G.N.Phillips,
and
M.P.Richards
(2009).
Structural analysis of fish versus mammalian hemoglobins: effect of the heme pocket environment on autooxidation and hemin loss.
|
| |
Proteins,
75,
217-230.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Janowski,
S.Panjikar,
A.N.Eddine,
S.H.Kaufmann,
and
M.S.Weiss
(2009).
Structural analysis reveals DNA binding properties of Rv2827c, a hypothetical protein from Mycobacterium tuberculosis.
|
| |
J Struct Funct Genomics,
10,
137-150.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Jin,
S.K.Singh,
S.Gu,
H.Furukawa,
A.I.Sobolevsky,
J.Zhou,
Y.Jin,
and
E.Gouaux
(2009).
Crystal structure and association behaviour of the GluR2 amino-terminal domain.
|
| |
EMBO J,
28,
1812-1823.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Potestio,
F.Pontiggia,
and
C.Micheletti
(2009).
Coarse-grained description of protein internal dynamics: an optimal strategy for decomposing proteins in rigid subunits.
|
| |
Biophys J,
96,
4993-5002.
|
 |
|
|
|
|
 |
S.D.Pegan,
K.Rukseree,
S.G.Franzblau,
and
A.D.Mesecar
(2009).
Structural basis for catalysis of a tetrameric class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis.
|
| |
J Mol Biol,
386,
1038-1053.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Aleksiev,
R.Potestio,
F.Pontiggia,
S.Cozzini,
and
C.Micheletti
(2009).
PiSQRD: a web server for decomposing proteins into quasi-rigid dynamical domains.
|
| |
Bioinformatics,
25,
2743-2744.
|
 |
|
|
|
|
 |
V.Majava,
and
P.Kursula
(2009).
Domain swapping and different oligomeric States for the complex between calmodulin and the calmodulin-binding domain of calcineurin a.
|
| |
PLoS ONE,
4,
e5402.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.Xu,
L.Yi,
Y.Feng,
L.Chen,
and
J.Liu
(2009).
Structural insight into the activation mechanism of human pancreatic prophospholipase A2.
|
| |
J Biol Chem,
284,
16659-16666.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.Min,
R.Akella,
H.He,
J.M.Humphreys,
S.E.Tsutakawa,
S.J.Lee,
J.A.Tainer,
M.H.Cobb,
and
E.J.Goldsmith
(2009).
The structure of the MAP2K MEK6 reveals an autoinhibitory dimer.
|
| |
Structure,
17,
96.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Qi,
M.C.Spong,
K.Nam,
A.Banerjee,
S.Jiralerspong,
M.Karplus,
and
G.L.Verdine
(2009).
Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme.
|
| |
Nature,
462,
762-766.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.J.Plested,
R.Vijayan,
P.C.Biggin,
and
M.L.Mayer
(2008).
Molecular basis of kainate receptor modulation by sodium.
|
| |
Neuron,
58,
720-735.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.K.Sendamarai,
R.S.Ohgami,
M.D.Fleming,
and
C.M.Lawrence
(2008).
Structure of the membrane proximal oxidoreductase domain of human Steap3, the dominant ferrireductase of the erythroid transferrin cycle.
|
| |
Proc Natl Acad Sci U S A,
105,
7410-7415.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Krumm,
X.Meng,
Y.Li,
Y.Xiang,
and
J.Deng
(2008).
Structural basis for antagonism of human interleukin 18 by poxvirus interleukin 18-binding protein.
|
| |
Proc Natl Acad Sci U S A,
105,
20711-20715.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.Nocek,
R.Mulligan,
M.Bargassa,
F.Collart,
and
A.Joachimiak
(2008).
Crystal structure of aminopeptidase N from human pathogen Neisseria meningitidis.
|
| |
Proteins,
70,
273-279.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.T.Kelly,
A.J.McCoy,
K.Späte,
S.E.Miller,
P.R.Evans,
S.Höning,
and
D.J.Owen
(2008).
A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex.
|
| |
Nature,
456,
976-979.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.T.Kelly,
A.J.McCoy,
K.Späte,
S.E.Miller,
P.R.Evans,
S.Höning,
and
D.J.Owen
(2008).
A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex.
|
| |
Nature,
456,
976-979.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.G.Roessler,
B.M.Hall,
W.J.Anderson,
W.M.Ingram,
S.A.Roberts,
W.R.Montfort,
and
M.H.Cordes
(2008).
Transitive homology-guided structural studies lead to discovery of Cro proteins with 40% sequence identity but different folds.
|
| |
Proc Natl Acad Sci U S A,
105,
2343-2348.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Manzano,
C.Contreras-Martel,
L.El Mortaji,
T.Izoré,
D.Fenel,
T.Vernet,
G.Schoehn,
A.M.Di Guilmi,
and
A.Dessen
(2008).
Sortase-mediated pilus fiber biogenesis in Streptococcus pneumoniae.
|
| |
Structure,
16,
1838-1848.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Zhang,
E.Bitto,
R.D.Goff,
S.Singh,
C.A.Bingman,
B.R.Griffith,
C.Albermann,
G.N.Phillips,
and
J.S.Thorson
(2008).
Biochemical and structural insights of the early glycosylation steps in calicheamicin biosynthesis.
|
| |
Chem Biol,
15,
842-853.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Fox,
I.Le Trong,
P.Rajagopal,
P.S.Brzovic,
R.E.Stenkamp,
and
R.E.Klevit
(2008).
Crystal structure of the BARD1 ankyrin repeat domain and its functional consequences.
|
| |
J Biol Chem,
283,
21179-21186.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Shi,
V.Sagar,
Z.Jin,
X.Yu,
L.Caldovic,
H.Morizono,
N.M.Allewell,
and
M.Tuchman
(2008).
The crystal structure of N-acetyl-L-glutamate synthase from Neisseria gonorrhoeae provides insights into mechanisms of catalysis and regulation.
|
| |
J Biol Chem,
283,
7176-7184.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Veesler,
S.Blangy,
C.Cambillau,
and
G.Sciara
(2008).
There is a baby in the bath water: AcrB contamination is a major problem in membrane-protein crystallization.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
880-885.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.A.Merritt,
M.Holmes,
F.S.Buckner,
W.C.Van Voorhis,
E.Quartly,
E.M.Phizicky,
A.Lauricella,
J.Luft,
G.DeTitta,
H.Neely,
F.Zucker,
and
W.G.Hol
(2008).
Structure of a Trypanosoma brucei alpha/beta-hydrolase fold protein with unknown function.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
474-478.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Bitto,
Y.Huang,
C.A.Bingman,
S.Singh,
J.S.Thorson,
and
G.N.Phillips
(2008).
The structure of flavin-dependent tryptophan 7-halogenase RebH.
|
| |
Proteins,
70,
289-293.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.Pontiggia,
A.Zen,
and
C.Micheletti
(2008).
Small- and large-scale conformational changes of adenylate kinase: a molecular dynamics study of the subdomain motion and mechanics.
|
| |
Biophys J,
95,
5901-5912.
|
 |
|
|
|
|
 |
H.L.Schubert,
J.D.Phillips,
A.Heroux,
and
C.P.Hill
(2008).
Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
|
| |
Biochemistry,
47,
8648-8655.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.L.Schubert,
R.S.Rose,
H.K.Leech,
A.A.Brindley,
C.P.Hill,
S.E.Rigby,
and
M.J.Warren
(2008).
Structure and function of SirC from Bacillus megaterium: a metal-binding precorrin-2 dehydrogenase.
|
| |
Biochem J,
415,
257-263.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.A.Potter,
R.E.Randall,
and
G.L.Taylor
(2008).
Crystal structure of human IPS-1/MAVS/VISA/Cardif caspase activation recruitment domain.
|
| |
BMC Struct Biol,
8,
11.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Groppe,
C.S.Hinck,
P.Samavarchi-Tehrani,
C.Zubieta,
J.P.Schuermann,
A.B.Taylor,
P.M.Schwarz,
J.L.Wrana,
and
A.P.Hinck
(2008).
Cooperative assembly of TGF-beta superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding.
|
| |
Mol Cell,
29,
157-168.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Guhaniyogi,
T.Wu,
S.S.Patel,
and
A.M.Stock
(2008).
Interaction of CheY with the C-terminal peptide of CheZ.
|
| |
J Bacteriol,
190,
1419-1428.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.T.Barglow,
K.S.Saikatendu,
M.H.Bracey,
R.Huey,
G.M.Morris,
A.J.Olson,
R.C.Stevens,
and
B.F.Cravatt
(2008).
Functional proteomic and structural insights into molecular recognition in the nitrilase family enzymes.
|
| |
Biochemistry,
47,
13514-13523.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.V.Korotkov,
and
W.G.Hol
(2008).
Structure of the GspK-GspI-GspJ complex from the enterotoxigenic Escherichia coli type 2 secretion system.
|
| |
Nat Struct Mol Biol,
15,
462-468.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.C.Pearce,
C.J.Morton,
S.C.Feil,
G.Hansen,
J.J.Adams,
M.W.Parker,
and
S.P.Bottomley
(2008).
Preventing serpin aggregation: the molecular mechanism of citrate action upon antitrypsin unfolding.
|
| |
Protein Sci,
17,
2127-2133.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.E.Yanez,
K.V.Korotkov,
J.Abendroth,
and
W.G.Hol
(2008).
The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the type 2 secretion system of Vibrio vulnificus.
|
| |
J Mol Biol,
375,
471-486.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.E.Yanez,
K.V.Korotkov,
J.Abendroth,
and
W.G.Hol
(2008).
Structure of the minor pseudopilin EpsH from the Type 2 secretion system of Vibrio cholerae.
|
| |
J Mol Biol,
377,
91.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Hirsch,
and
M.Habeck
(2008).
Mixture models for protein structure ensembles.
|
| |
Bioinformatics,
24,
2184-2192.
|
 |
|
|
|
|
 |
M.Monné,
L.Han,
T.Schwend,
S.Burendahl,
and
L.Jovine
(2008).
Crystal structure of the ZP-N domain of ZP3 reveals the core fold of animal egg coats.
|
| |
Nature,
456,
653-657.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Resch,
H.Striegl,
E.M.Henssler,
M.Sevvana,
C.Egerer-Sieber,
E.Schiltz,
W.Hillen,
and
Y.A.Muller
(2008).
A protein functional leap: how a single mutation reverses the function of the transcription regulator TetR.
|
| |
Nucleic Acids Res,
36,
4390-4401.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Rushe,
L.Silvian,
S.Bixler,
L.L.Chen,
A.Cheung,
S.Bowes,
H.Cuervo,
S.Berkowitz,
T.Zheng,
K.Guckian,
M.Pellegrini,
and
A.Lugovskoy
(2008).
Structure of a NEMO/IKK-associating domain reveals architecture of the interaction site.
|
| |
Structure,
16,
798-808.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.S.Murray,
R.P.Holmes,
and
W.T.Lowther
(2008).
Active site and loop 4 movements within human glycolate oxidase: implications for substrate specificity and drug design.
|
| |
Biochemistry,
47,
2439-2449.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.W.Vetting,
J.C.Errey,
and
J.S.Blanchard
(2008).
Rv0802c from Mycobacterium tuberculosis: the first structure of a succinyltransferase with the GNAT fold.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
978-985.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.X.Mori,
C.W.Vander Kooi,
D.J.Leahy,
and
D.T.Yue
(2008).
Crystal structure of the CaV2 IQ domain in complex with Ca2+/calmodulin: high-resolution mechanistic implications for channel regulation by Ca2+.
|
| |
Structure,
16,
607-620.
|
 |
|
|
|
|
 |
N.Handa,
E.Mizohata,
S.Kishishita,
M.Toyama,
S.Morita,
T.Uchikubo-Kamo,
R.Akasaka,
K.Omori,
J.Kotera,
T.Terada,
M.Shirouzu,
and
S.Yokoyama
(2008).
Crystal structure of the GAF-B domain from human phosphodiesterase 10A complexed with its ligand, cAMP.
|
| |
J Biol Chem,
283,
19657-19664.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Schrader,
P.Stelter,
D.Flemming,
R.Kunze,
E.Hurt,
and
I.R.Vetter
(2008).
Structural basis of the nic96 subcomplex organization in the nuclear pore channel.
|
| |
Mol Cell,
29,
46-55.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Filippakopoulos,
M.Kofler,
O.Hantschel,
G.D.Gish,
F.Grebien,
E.Salah,
P.Neudecker,
L.E.Kay,
B.E.Turk,
G.Superti-Furga,
T.Pawson,
and
S.Knapp
(2008).
Structural coupling of SH2-kinase domains links Fes and Abl substrate recognition and kinase activation.
|
| |
Cell,
134,
793-803.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Q.Zhai,
R.D.Fisher,
H.Y.Chung,
D.G.Myszka,
W.I.Sundquist,
and
C.P.Hill
(2008).
Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV.
|
| |
Nat Struct Mol Biol,
15,
43-49.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.P.Grant,
N.J.Marshall,
J.C.Yang,
M.B.Fasken,
S.M.Kelly,
M.T.Harreman,
D.Neuhaus,
A.H.Corbett,
and
M.Stewart
(2008).
Structure of the N-terminal Mlp1-binding domain of the Saccharomyces cerevisiae mRNA-binding protein, Nab2.
|
| |
J Mol Biol,
376,
1048-1059.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Stamler,
H.T.Keutmann,
Y.Sidis,
C.Kattamuri,
A.Schneyer,
and
T.B.Thompson
(2008).
The Structure of FSTL3{middle dot}Activin A Complex: DIFFERENTIAL BINDING OF N-TERMINAL DOMAINS INFLUENCES FOLLISTATIN-TYPE ANTAGONIST SPECIFICITY.
|
| |
J Biol Chem,
283,
32831-32838.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.C.Flores,
K.S.Keating,
J.Painter,
F.Morcos,
K.Nguyen,
E.A.Merritt,
L.A.Kuhn,
and
M.B.Gerstein
(2008).
HingeMaster: normal mode hinge prediction approach and integration of complementary predictors.
|
| |
Proteins,
73,
299-319.
|
 |
|
|
|
|
 |
S.O.Nilsson Lill,
J.Gao,
and
G.L.Waldrop
(2008).
Molecular dynamics simulations of biotin carboxylase.
|
| |
J Phys Chem B,
112,
3149-3156.
|
 |
|
|
|
|
 |
T.A.Springer,
J.Zhu,
and
T.Xiao
(2008).
Structural basis for distinctive recognition of fibrinogen gammaC peptide by the platelet integrin alphaIIbbeta3.
|
| |
J Cell Biol,
182,
791-800.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.J.Zhang,
Y.X.He,
Z.Yang,
J.Yu,
Y.Chen,
and
C.Z.Zhou
(2008).
Crystal structure of glutathione-dependent phospholipid peroxidase Hyr1 from the yeast Saccharomyces cerevisiae.
|
| |
Proteins,
73,
1058-1062.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Yao,
C.B.Harrison,
P.L.Freddolino,
K.Schulten,
and
M.L.Mayer
(2008).
Molecular mechanism of ligand recognition by NR3 subtype glutamate receptors.
|
| |
EMBO J,
27,
2158-2170.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.K.Poon,
X.Chen,
M.Lu,
N.K.Vyas,
F.A.Quiocho,
Q.Wang,
and
J.Ma
(2007).
Normal mode refinement of anisotropic thermal parameters for a supramolecular complex at 3.42-A crystallographic resolution.
|
| |
Proc Natl Acad Sci U S A,
104,
7869-7874.
|
 |
|
|
|
|
 |
C.E.Christensen,
B.B.Kragelund,
P.von Wettstein-Knowles,
and
A.Henriksen
(2007).
Structure of the human beta-ketoacyl [ACP] synthase from the mitochondrial type II fatty acid synthase.
|
| |
Protein Sci,
16,
261-272.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.W.Vander Kooi,
M.A.Jusino,
B.Perman,
D.B.Neau,
H.D.Bellamy,
and
D.J.Leahy
(2007).
Structural basis for ligand and heparin binding to neuropilin B domains.
|
| |
Proc Natl Acad Sci U S A,
104,
6152-6157.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Eyal,
C.Chennubhotla,
L.W.Yang,
and
I.Bahar
(2007).
Anisotropic fluctuations of amino acids in protein structures: insights from X-ray crystallography and elastic network models.
|
| |
Bioinformatics,
23,
i175-i184.
|
 |
|
|
|
|
 |
G.Zocher,
U.Wiesand,
and
G.E.Schulz
(2007).
High resolution structure and catalysis of O-acetylserine sulfhydrylase isozyme B from Escherichia coli.
|
| |
FEBS J,
274,
5382-5389.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.M.Kim,
B.S.Park,
J.I.Kim,
S.E.Kim,
J.Lee,
S.C.Oh,
P.Enkhbayar,
N.Matsushima,
H.Lee,
O.J.Yoo,
and
J.O.Lee
(2007).
Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran.
|
| |
Cell,
130,
906-917.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.P.Fabrichny,
P.Leone,
G.Sulzenbacher,
D.Comoletti,
M.T.Miller,
P.Taylor,
Y.Bourne,
and
P.Marchot
(2007).
Structural analysis of the synaptic protein neuroligin and its beta-neurexin complex: determinants for folding and cell adhesion.
|
| |
Neuron,
56,
979-991.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.A.Garnett,
S.Baumberg,
P.G.Stockley,
and
S.E.Phillips
(2007).
Structure of the C-terminal effector-binding domain of AhrC bound to its corepressor L-arginine.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
918-921.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.C.Grigg,
C.L.Vermeiren,
D.E.Heinrichs,
and
M.E.Murphy
(2007).
Heme coordination by Staphylococcus aureus IsdE.
|
| |
J Biol Chem,
282,
28815-28822.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.G.McCoy,
E.Bitto,
C.A.Bingman,
G.E.Wesenberg,
R.M.Bannen,
D.A.Kondrashov,
and
G.N.Phillips
(2007).
Structure and dynamics of UDP-glucose pyrophosphorylase from Arabidopsis thaliana with bound UDP-glucose and UTP.
|
| |
J Mol Biol,
366,
830-841.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.J.Warren,
T.J.Pohlhaus,
A.Changela,
R.R.Iyer,
P.L.Modrich,
and
L.S.Beese
(2007).
Structure of the human MutSalpha DNA lesion recognition complex.
|
| |
Mol Cell,
26,
579-592.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Lee,
A.R.Feldman,
B.Delmas,
and
M.Paetzel
(2007).
Crystal structure of the VP4 protease from infectious pancreatic necrosis virus reveals the acyl-enzyme complex for an intermolecular self-cleavage reaction.
|
| |
J Biol Chem,
282,
24928-24937.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.I.Llarrull,
S.M.Fabiane,
J.M.Kowalski,
B.Bennett,
B.J.Sutton,
and
A.J.Vila
(2007).
Asp-120 locates Zn2 for optimal metallo-beta-lactamase activity.
|
| |
J Biol Chem,
282,
18276-18285.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.L.Mendillo,
C.D.Putnam,
and
R.D.Kolodner
(2007).
Escherichia coli MutS tetramerization domain structure reveals that stable dimers but not tetramers are essential for DNA mismatch repair in vivo.
|
| |
J Biol Chem,
282,
16345-16354.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Schnell,
W.Oehlmann,
M.Singh,
and
G.Schneider
(2007).
Structural insights into catalysis and inhibition of O-acetylserine sulfhydrylase from Mycobacterium tuberculosis. Crystal structures of the enzyme alpha-aminoacrylate intermediate and an enzyme-inhibitor complex.
|
| |
J Biol Chem,
282,
23473-23481.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.C.Flores,
L.J.Lu,
J.Yang,
N.Carriero,
and
M.B.Gerstein
(2007).
Hinge Atlas: relating protein sequence to sites of structural flexibility.
|
| |
BMC Bioinformatics,
8,
167.
|
 |
|
|
|
|
 |
S.Meyer,
S.Savaresi,
I.C.Forster,
and
R.Dutzler
(2007).
Nucleotide recognition by the cytoplasmic domain of the human chloride transporter ClC-5.
|
| |
Nat Struct Mol Biol,
14,
60-67.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Mouilleron,
and
B.Golinelli-Pimpaneau
(2007).
Domain motions of glucosamine-6P synthase: comparison of the anisotropic displacements in the crystals and the catalytic hinge-bending rotation.
|
| |
Protein Sci,
16,
485-493.
|
 |
|
|
|
|
 |
T.W.Geders,
L.Gu,
J.C.Mowers,
H.Liu,
W.H.Gerwick,
K.Håkansson,
D.H.Sherman,
and
J.L.Smith
(2007).
Crystal structure of the ECH2 catalytic domain of CurF from Lyngbya majuscula. Insights into a decarboxylase involved in polyketide chain beta-branching.
|
| |
J Biol Chem,
282,
35954-35963.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.A.Stanley,
N.V.Pursiainen,
E.F.Garman,
A.H.Juffer,
M.Wilmanns,
and
P.Kursula
(2007).
A previously unobserved conformation for the human Pex5p receptor suggests roles for intrinsic flexibility and rigid domain motions in ligand binding.
|
| |
BMC Struct Biol,
7,
24.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.Cheng,
I.Ivanov,
H.Wang,
S.M.Sine,
and
J.A.McCammon
(2007).
Nanosecond-timescale conformational dynamics of the human alpha7 nicotinic acetylcholine receptor.
|
| |
Biophys J,
93,
2622-2634.
|
 |
|
|
|
|
 |
X.Tan,
L.I.Calderon-Villalobos,
M.Sharon,
C.Zheng,
C.V.Robinson,
M.Estelle,
and
N.Zheng
(2007).
Mechanism of auxin perception by the TIR1 ubiquitin ligase.
|
| |
Nature,
446,
640-645.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.C.Weston,
P.Schuck,
A.Ghosal,
C.Rosenmund,
and
M.L.Mayer
(2006).
Conformational restriction blocks glutamate receptor desensitization.
|
| |
Nat Struct Mol Biol,
13,
1120-1127.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

