 |
PDBsum entry 3crm

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.5.1.75
- tRNA dimethylallyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
adenosine37 in tRNA + dimethylallyl diphosphate = N6- dimethylallyladenosine37 in tRNA + diphosphate
|
 |
 |
 |
 |
 |
adenosine(37) in tRNA
|
+
|
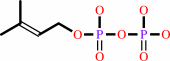 dimethylallyl diphosphate
dimethylallyl diphosphate
|
=
|
N(6)- dimethylallyladenosine(37) in tRNA
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
367:872-881
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of tRNA dimethylallyltransferase: RNA modification through a channel.
|
|
W.Xie,
C.Zhou,
R.H.Huang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Dimethylallyltransferase (DMATase) transfers a five-carbon isoprenoid moiety
from dimethylallyl pyrophosphate (DMAPP) to the amino group of adenosine at
position 37 of certain tRNAs. Reported here are the crystal structures of
Pseudomonas aeruginosa DMATase alone and in complex with pyrophosphate at 1.9 A
resolution. Surprisingly, the enzyme possesses a central channel spanning the
entire width of the enzyme. Both the accepting substrate tRNA and the donating
substrate DMAPP appear to enter the channel from opposite sides in an ordered
sequence, with tRNA first and DMAPP second, and the RNA modification reaction
occurs in the middle of the channel once the two substrates have met. The
structure of DMATase is homologous to a class of small soluble kinases involved
in biosynthesis of nucleotide precursors for nucleic acids, indicating its
possibly evolutionary origin. Furthermore, specific recognition of the
pyrophosphate by a conserved loop in DMATase, similar to the P-loop commonly
seen in diverse nucleotide-binding proteins, demonstrates that DMATase is
structurally and mechanistically distinct from farnesyltransferase, another
family of prenyltransferases involved in protein modification.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4. Stereo view of C^α superposition of the DMATase
structure with the three top scores from Dali structural
homology search. DMATase is in blue, S. cerevisiae GMP kinase
(PDB entry 1GKY, Z core = 12.1, rmsd = 2.9 Å) is in red,
human UMP/CMP kinase (PDB entry 1TEV, Z core = 11.8, rmsd = 3.3
Å) is in magenta, and Leishmania major CMP kinase (PDB
entry 1Y63, Z core = 10.9, rmsd = 3.2 Å) is in yellow.
Figure 4. Stereo view of C^α superposition of the DMATase
structure with the three top scores from Dali structural
homology search. DMATase is in blue, S. cerevisiae GMP kinase
(PDB entry 1GKY, Z core = 12.1, rmsd = 2.9 Å) is in red,
human UMP/CMP kinase (PDB entry 1TEV, Z core = 11.8, rmsd = 3.3
Å) is in magenta, and Leishmania major CMP kinase (PDB
entry 1Y63, Z core = 10.9, rmsd = 3.2 Å) is in yellow.
|
 |
Figure 7.
Figure 7. Proposed mechanism of DMATase-catalyzed reaction. The
tRNA substrate binds to the right side of channel and the base
of A37 flips into the channel (step 1). DMAPP enters the
channel from the left side and is stabilized by interacting with
the P-loop as well as by coordinating with a Mg^2+ (step 2).
Nucleophilic attack of the amino group in A37 on the DMA moiety
of DMAPP results in the DMA moiety to be transferred from DMAPP
to the A37 of tRNA, made possible by activation of the carbon
atom directly linked to the pyrophosphate in DMAPP by the
side-chains of T14 and R223, as well as deprotonation of the
amino group in A37 with the side-chain of D37 (step 3).
Figure 7. Proposed mechanism of DMATase-catalyzed reaction. The
tRNA substrate binds to the right side of channel and the base
of A37 flips into the channel (step 1). DMAPP enters the channel
from the left side and is stabilized by interacting with the
P-loop as well as by coordinating with a Mg^2+ (step 2).
Nucleophilic attack of the amino group in A37 on the DMA moiety
of DMAPP results in the DMA moiety to be transferred from DMAPP
to the A37 of tRNA, made possible by activation of the carbon
atom directly linked to the pyrophosphate in DMAPP by the
side-chains of T14 and R223, as well as deprotonation of the
amino group in A37 with the side-chain of D37 (step 3).
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Elsevier:
J Mol Biol
(2007,
367,
872-881)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.M.Chu,
T.P.Ko,
and
A.H.Wang
(2010).
Crystal structure and substrate specificity of plant adenylate isopentenyltransferase from Humulus lupulus: distinctive binding affinity for purine and pyrimidine nucleotides.
|
| |
Nucleic Acids Res,
38,
1738-1748.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Seif,
and
B.M.Hallberg
(2009).
RNA-Protein Mutually Induced Fit: STRUCTURE OF ESCHERICHIA COLI ISOPENTENYL-tRNA TRANSFERASE IN COMPLEX WITH tRNA(Phe).
|
| |
J Biol Chem,
284,
6600-6604.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.L.Andreassi,
M.W.Vetting,
P.W.Bilder,
S.L.Roderick,
and
T.S.Leyh
(2009).
Structure of the ternary complex of phosphomevalonate kinase: the enzyme and its family.
|
| |
Biochemistry,
48,
6461-6468.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Chimnaronk,
F.Forouhar,
J.Sakai,
M.Yao,
C.M.Tron,
M.Atta,
M.Fontecave,
J.F.Hunt,
and
I.Tanaka
(2009).
Snapshots of dynamics in synthesizing N(6)-isopentenyladenosine at the tRNA anticodon.
|
| |
Biochemistry,
48,
5057-5065.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Zhou,
and
R.H.Huang
(2008).
Crystallographic snapshots of eukaryotic dimethylallyltransferase acting on tRNA: insight into tRNA recognition and reaction mechanism.
|
| |
Proc Natl Acad Sci U S A,
105,
16142-16147.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

