 |
PDBsum entry 3chp

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.3.2.6
- leukotriene-A4 hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
leukotriene A4 + H2O = leukotriene B4
|
 |
 |
 |
 |
 |
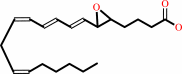 leukotriene A4
leukotriene A4
|
+
|
H2O
|
=
|
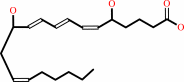 leukotriene B4
leukotriene B4
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.4.11.4
- tripeptide aminopeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Release of a N-terminal residue from a tripeptide.
|
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Bioorg Med Chem Lett
16:4963-4983
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
|
|
T.A.Kirkland,
M.Adler,
J.G.Bauman,
M.Chen,
J.Z.Haeggström,
B.King,
M.J.Kochanny,
A.M.Liang,
L.Mendoza,
G.B.Phillips,
M.Thunnissen,
L.Trinh,
M.Whitlow,
B.Ye,
H.Ye,
J.Parkinson,
W.J.Guilford.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Leukotriene B(4) (LTB(4)) is a potent pro-inflammatory mediator that has been
implicated in the pathogenesis of multiple diseases, including psoriasis,
inflammatory bowel disease, multiple sclerosis and asthma. As a method to
decrease the level of LTB(4) and possibly identify novel treatments, inhibitors
of the LTB(4) biosynthetic enzyme, leukotriene A(4) hydrolase (LTA(4)-h), have
been explored. Here we describe the discovery of a potent inhibitor of LTA(4)-h,
arylamide of glutamic acid 4f, starting from the corresponding glycinamide 2.
Analogs of 4f are then described, focusing on compounds that are both active and
stable in whole blood. This effort culminated in the identification of amino
alcohol 12a and amino ester 6b which meet these criteria.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Thangapandian,
S.John,
S.Sakkiah,
and
K.W.Lee
(2011).
Pharmacophore-based virtual screening and Bayesian model for the identification of potential human leukotriene A4 hydrolase inhibitors.
|
| |
Eur J Med Chem,
46,
1593-1603.
|
 |
|
|
|
|
 |
A.Mirshafiey,
and
F.Jadidi-Niaragh
(2010).
Immunopharmacological role of the leukotriene receptor antagonists and inhibitors of leukotrienes generating enzymes in multiple sclerosis.
|
| |
Immunopharmacol Immunotoxicol,
32,
219-227.
|
 |
|
|
|
|
 |
D.R.Davies,
B.Mamat,
O.T.Magnusson,
J.Christensen,
M.H.Haraldsson,
R.Mishra,
B.Pease,
E.Hansen,
J.Singh,
D.Zembower,
H.Kim,
A.S.Kiselyov,
A.B.Burgin,
M.E.Gurney,
and
L.J.Stewart
(2009).
Discovery of leukotriene A4 hydrolase inhibitors using metabolomics biased fragment crystallography.
|
| |
J Med Chem,
52,
4694-4715.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

