 |
PDBsum entry 3b8d

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.2.13
- fructose-bisphosphate aldolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 1,6-bisphosphate = D-glyceraldehyde 3-phosphate + dihydroxyacetone phosphate
|
 |
 |
 |
 |
 |
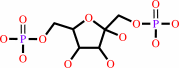 beta-D-fructose 1,6-bisphosphate
beta-D-fructose 1,6-bisphosphate
|
=
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
+
|
 dihydroxyacetone phosphate
dihydroxyacetone phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
277:9474-9483
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
A conserved glutamate residue exhibits multifunctional catalytic roles in D-fructose-1,6-bisphosphate aldolases.
|
|
A.Maurady,
A.Zdanov,
D.de Moissac,
D.Beaudry,
J.Sygusch.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The aldolase catalytic cycle consists of a number of proton transfers that
interconvert covalent enzyme intermediates. Glu-187 is a conserved amino acid
that is located in the mammalian fructose-1,6-bisphosphate aldolase active site.
Its central location, within hydrogen bonding distance of three other conserved
active site residues: Lys-146, Glu-189, and Schiff base-forming Lys-229, makes
it an ideal candidate for mediating proton transfers. Point mutations,
Glu-187--> Gln, Ala, which would inhibit proton transfers significantly,
compromise activity. Trapping of enzymatic intermediates in Glu-187 mutants
defines a proton transfer role for Glu-187 in substrate cleavage and Schiff base
formation. Structural data show that loss of Glu-187 negative charge results in
hydrogen bond formation between Lys-146 and Lys-229 consistent with a basic
pK(a) for Lys-229 in native enzyme and supporting nucleophilic activation of
Lys-229 by Glu-187 during Schiff base formation. The crystal structures also
substantiate Glu-187 and Glu-189 as present in ionized form in native enzyme,
compatible with their role of catalyzing proton exchange with solvent as
indicated from solvent isotope effects. The proton exchange mechanism ensures
Glu-187 basicity throughout the catalytic cycle requisite for mediating proton
transfer and electrostatic stabilization of ketamine intermediates. Glutamate
general base catalysis is a recurrent evolutionary feature of Schiff
base0forming aldolases.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. Stereoview of electron density showing Gln-187,
Glu-189, and Arg-148 residues in the active site of the E187Q
mutant structure. The mutant structure is shown superimposed
with equivalent residues in the native enzyme (dark green).
Gln-187 donates a hydrogen bond to Glu-189 in E187Q whereas
Arg-148 makes additional hydrogen bonds with Glu-189 in E187Q
not observed in the native structure. Wat-1376 makes a hydrogen
bond to Glu-189 whereas Wat-1647 interacts with Glu-189 and
Wat-1856. Electron density shown correspond to a 2F[o]  F[c] omit
map of residue Gln-187 and contoured at the 1 F[c] omit
map of residue Gln-187 and contoured at the 1  level. level.
|
 |
Figure 4.
Fig. 4. Stereoview of electron density showing
superposition of Lys-146, Gln-187, Lys-229, and Leu-270 in E187Q
mutant with equivalent residues in the native enzyme (dark
green). The hydrogen bond between lysine residues requires that
one lysine residue acts as hydrogen bond acceptor. Glu-187 in
the native structure is situated within hydrogen bonding
distance between the two lysine residues. Wat-8272 makes
hydrogen bonds to Lys-146 and Wat-8338 whereas Leu-270 makes
close contact with Lys-229. Electron densities shown correspond
to a 2F[o]  F[c] omit
map of residues Lys-146 and Lys-229 and contoured at the 1 F[c] omit
map of residues Lys-146 and Lys-229 and contoured at the 1  level. level.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2002,
277,
9474-9483)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.W.Song,
J.G.Lee,
H.S.Youn,
S.H.Eom,
and
d.o. .H.Kim
(2011).
Ryanodine receptor assembly: A novel systems biology approach to 3D mapping.
|
| |
Prog Biophys Mol Biol,
105,
145-161.
|
 |
|
|
|
|
 |
M.Capela,
N.J.Mosey,
L.Xing,
R.Wang,
and
A.Petitjean
(2011).
Amine exchange in formamidines: an experimental and theoretical study.
|
| |
Chemistry,
17,
4598-4612.
|
 |
|
|
|
|
 |
Y.Sato,
and
M.Nishida
(2009).
Electric charge divergence in proteins: insights into the evolution of their three-dimensional properties.
|
| |
Gene,
441,
3.
|
 |
|
|
|
|
 |
C.A.Buscaglia,
W.G.Hol,
V.Nussenzweig,
and
T.Cardozo
(2007).
Modeling the interaction between aldolase and the thrombospondin-related anonymous protein, a key connection of the malaria parasite invasion machinery.
|
| |
Proteins,
66,
528-537.
|
 |
|
|
|
|
 |
J.A.Pezza,
J.D.Stopa,
E.M.Brunyak,
K.N.Allen,
and
D.R.Tolan
(2007).
Thermodynamic analysis shows conformational coupling and dynamics confer substrate specificity in fructose-1,6-bisphosphate aldolase.
|
| |
Biochemistry,
46,
13010-13018.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

