 |
PDBsum entry 3zo7
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of clcfe27a with substrate
|
|
Structure:
|
 |
5-chloromuconolactone dehalogenase. Chain: a, b, c, d, e, f, g, h, i, j. Synonym: chlormuconolactone dehalogenase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Rhodococcus opacus. Organism_taxid: 37919. Strain: 1cp. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.22Å
|
R-factor:
|
0.187
|
R-free:
|
0.232
|
|
|
Authors:
|
 |
C.Roth,J.A.D.Groening,S.R.Kaschabek,M.Schloemann,N.Straeter
|
|
Key ref:
|
 |
C.Roth
et al.
(2013).
Crystal structure and catalytic mechanism of chloromuconolactone dehalogenase ClcF from Rhodococcus opacus 1CP.
Mol Microbiol,
88,
254-267.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
20-Feb-13
|
Release date:
|
06-Mar-13
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q8G9L0
(Q8G9L0_RHOOP) -
Muconolactone Delta-isomerase from Rhodococcus opacus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
94 a.a.
92 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.3.3.4
- muconolactone Delta-isomerase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Benzoate Metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-muconolactone = (4,5-dihydro-5-oxofuran-2-yl)-acetate
|
 |
 |
 |
 |
 |
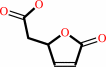 (+)-muconolactone
(+)-muconolactone
|
=
|
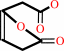 5-oxo-4,5-dihydrofuran-2-acetate
5-oxo-4,5-dihydrofuran-2-acetate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Mol Microbiol
88:254-267
(2013)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure and catalytic mechanism of chloromuconolactone dehalogenase ClcF from Rhodococcus opacus 1CP.
|
|
C.Roth,
J.A.Gröning,
S.R.Kaschabek,
M.Schlömann,
N.Sträter.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The actinobacterium Rhodococcus opacus 1CP possesses a so far unique variant of
the modified 3-oxoadipate pathway for 3-chlorocatechol degradation. One
important feature is the novel dehalogenase ClcF, which converts
(4R,5S)-5-chloromuconolactone to E-dienelactone. ClcF is related to
muconolactone isomerase (MLI, EC 5.3.3.4). The enzyme has a ferredoxin-type fold
and forms a homodecamer of 52-symmetry, typical for the MLI family. The active
site is formed by residues from two monomers. The complex structure of an E27A
variant with bound substrate in conjunction with mutational studies indicate
that E27 acts as the proton acceptor in a univalent single-base
syn-dehydrohalogenation mechanism. Despite the evolutionary specialization of
ClcF, the conserved active-site structures suggest that the proposed mechanism
is representative for the MLI family. Furthermore, ClcF represents a novel type
of dehalogenase based on an isomerase scaffold.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

