 |
PDBsum entry 3vml

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
3vml
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Chimera 3-isopropylmalate dehydrogenase between shewanella oneidensis mr-1 (o) and shewanella benthica db21 mt-2 (m) from n-terminal: 20% o middle 70% m residual 10% o
|
|
Structure:
|
 |
3-isopropylmalate dehydrogenase. Chain: a. Synonym: 3-ipm-dh, beta-ipm dehydrogenase, imdh. Engineered: yes. Other_details: chimera protein of 3-isopropylmalate dehydrogenase from shewanella oneidensis mr-1 (unp q8e9n3 residues 2-68), 3- isopropylmalate dehydrogenase from shewanella benthica db21 mt-2 (unp d2yzl2 residues 69-328), and 3-isopropylmalate dehydrogenase from shewanella oneidensis mr-1 (unp q8e9n3 residues 329-364)
|
|
Source:
|
 |
Shewanella oneidensis, shewanella benthica. Organism_taxid: 211586, 43661. Strain: mr-1, db21 mt-2, mr-1. Gene: leub, so_4235. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.56Å
|
R-factor:
|
0.179
|
R-free:
|
0.209
|
|
|
Authors:
|
 |
T.Nagae,N.Watanabe
|
|
Key ref:
|
 |
Y.Hamajima
et al.
Structure analysis of chimeric 3-Isopropylmalate dehydrogenase between the obligate piezophile shewane benthica db21mt-2 and the nonpiezophile shewanella oneidensis mr-1.
To be published,
.
|
 |
|
Date:
|
 |
|
13-Dec-11
|
Release date:
|
26-Dec-12
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.85
- 3-isopropylmalate dehydrogenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Leucine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2R,3S)-3-isopropylmalate + NAD+ = 4-methyl-2-oxopentanoate + CO2 + NADH
|
 |
 |
 |
 |
 |
(2R,3S)-3-isopropylmalate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 NAD(+)
NAD(+)
|
=
|
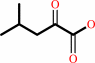 4-methyl-2-oxopentanoate
4-methyl-2-oxopentanoate
|
+
|
 CO2
CO2
|
+
|
 NADH
NADH
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
|

