 |
PDBsum entry 3nvt

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase/isomerase
|
PDB id
|
|

|

|
3nvt
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase/isomerase
|
 |
|
Title:
|
 |
1.95 angstrom crystal structure of a bifunctional 3-deoxy-7- phosphoheptulonate synthase/chorismate mutase (aroa) from listeria monocytogenes egd-e
|
|
Structure:
|
 |
3-deoxy-d-arabino-heptulosonate 7-phosphate synthase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Listeria monocytogenes. Organism_taxid: 169963. Strain: egd-e. Gene: aroa, lmo1600. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.95Å
|
R-factor:
|
0.156
|
R-free:
|
0.198
|
|
|
Authors:
|
 |
A.S.Halavaty,S.H.Light,G.Minasov,L.Shuvalova,K.Kwon,W.F.Anderson, Center For Structural Genomics Of Infectious Diseases (Csgid)
|
|
Key ref:
|
 |
S.H.Light
et al.
(2012).
Structural analysis of a 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase with an N-terminal chorismate mutase-like regulatory domain.
Protein Sci,
21,
887-895.
PubMed id:
|
 |
|
Date:
|
 |
|
08-Jul-10
|
Release date:
|
28-Jul-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chains A, B:
E.C.2.5.1.54
- 3-deoxy-7-phosphoheptulonate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Shikimate and Chorismate Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-erythrose 4-phosphate + phosphoenolpyruvate + H2O = 7-phospho-2- dehydro-3-deoxy-D-arabino-heptonate + phosphate
|
 |
 |
 |
 |
 |
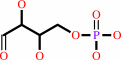 D-erythrose 4-phosphate
D-erythrose 4-phosphate
|
+
|
 phosphoenolpyruvate
phosphoenolpyruvate
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
=
|
7-phospho-2- dehydro-3-deoxy-D-arabino-heptonate
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B:
E.C.5.4.99.5
- chorismate mutase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
chorismate = prephenate
|
 |
 |
 |
 |
 |
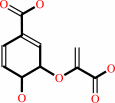 chorismate
chorismate
|
=
|
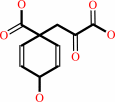 prephenate
prephenate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Protein Sci
21:887-895
(2012)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural analysis of a 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase with an N-terminal chorismate mutase-like regulatory domain.
|
|
S.H.Light,
A.S.Halavaty,
G.Minasov,
L.Shuvalova,
W.F.Anderson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

